Oxygen-induced alterations in the expression of chromatin modifying enzymes and the transcriptional regulation of imprinted genes
- PMID: 29339137
- PMCID: PMC6094953
- DOI: 10.1016/j.gep.2018.01.001
Oxygen-induced alterations in the expression of chromatin modifying enzymes and the transcriptional regulation of imprinted genes
Abstract
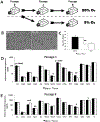
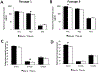
Embryo culture and assisted reproductive technologies have been associated with a disproportionately high number of epigenetic abnormalities in the resulting offspring. However, the mechanisms by which these techniques influence the epigenome remain poorly defined. In this study, we evaluated the capacity of oxygen concentration to influence the transcriptional control of a selection of key enzymes regulating chromatin structure. In mouse embryonic stem cells, oxygen concentrations modulated the transcriptional regulation of the TET family of enzymes, as well as the de novo methyltransferase Dnmt3a. These transcriptional changes were associated with alterations in the control of multiple imprinted genes, including H19, Igf2, Igf2r, and Peg3. Similarly, exposure of in vitro produced bovine embryos to atmospheric oxygen concentrations was associated with disruptions in the transcriptional regulation of TET1, TET3, and DNMT3a, along with the DNA methyltransferase co-factor HELLS. In addition, exposure to high oxygen was associated with alterations in the abundance of transcripts encoding members of the Polycomb repressor complex (EED and EZH2), the histone methyltransferase SETDB1 and multiple histone demethylases (KDM1A, KDM4B, and KDM4C). These disruptions were accompanied by a reduction in embryo viability and suppression of the pluripotency genes NANOG and SOX2. These experiments demonstrate that oxygen has the capacity to modulate the transcriptional control of chromatin modifying genes involved in the establishment and maintenance of both pluripotency and genomic imprinting.
Keywords: Assisted reproductive technologies; DNA methylation; DNMT; Developmental programming; Epigenetics; Genomic imprinting; Histone demethylase; Oxidative stress; TET.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
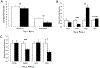



References
-
- Arnaud P, Feil R, 2005, Jun. Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res. Part C Embryo Today - Rev 75 (2), 81–97. - PubMed
-
- Bartolomei MS, Tilghman SM, 1997. Genomic imprinting in mammals. Annu. Rev. Genet 31 (1), 493–525. - PubMed
-
- Chen H-F, Kuo H-C, Chen W, Wu F-C, Yang Y-S, Ho H-N, 2009, Jan. A reduced oxygen tension (5%) is not beneficial for maintaining human embryonic stem cells in the undifferentiated state with short splitting intervals. Hum. Reprod 24 (1), 71–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

