Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer
- PMID: 29339376
- PMCID: PMC6047936
- DOI: 10.1158/2326-6066.CIR-17-0559
Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer
Abstract
The immune system exerts antitumor activity via T cell-dependent recognition of tumor-specific antigens. Although the number of tumor neopeptides-peptides derived from somatic mutations-often correlates with immune activity and survival, most classically defined high-affinity neopeptides (CDNs) are not immunogenic, and only rare CDNs have been linked to tumor rejection. Thus, the rules of tumor antigen recognition remain incompletely understood. Here, we analyzed neopeptides, immune activity, and clinical outcome from 6,324 patients across 27 tumor types. We characterized a class of "alternatively defined neopeptides" (ADNs), which are mutant peptides predicted to bind MHC (class I or II) with improved affinity relative to their nonmutated counterpart. ADNs are abundant and molecularly distinct from CDNs. The load of ADNs correlated with intratumoral T-cell responses and immune suppression, and ADNs were also strong predictors of patient survival across tumor types. These results expand the spectrum of mutation-derived tumor antigens with potential clinical relevance. Cancer Immunol Res; 6(3); 276-87. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures



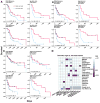
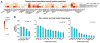
References
-
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

