Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway in Arabidopsis
- PMID: 29339505
- PMCID: PMC5798382
- DOI: 10.1073/pnas.1719497115
Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway in Arabidopsis
Abstract
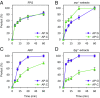
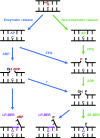
Abasic (apurinic/apyrimidinic, AP) sites in DNA arise from spontaneous base loss or by enzymatic removal during base excision repair. It is commonly accepted that both classes of AP site have analogous biochemical properties and are equivalent substrates for AP endonucleases and AP lyases, although the relative roles of these two types of enzymes are not well understood. We provide here genetic and biochemical evidence that, in Arabidopsis, AP sites generated by spontaneous loss of N7-methylguanine (N7-meG) are exclusively repaired through an AP endonuclease-independent pathway initiated by FPG, a bifunctional DNA glycosylase with AP lyase activity. Abasic site incision catalyzed by FPG generates a single-nucleotide gap with a 3'-phosphate terminus that is processed by the DNA 3'-phosphatase ZDP before repair is completed. We further show that the major AP endonuclease in Arabidopsis (ARP) incises AP sites generated by enzymatic N7-meG excision but, unexpectedly, not those resulting from spontaneous N7-meG loss. These findings, which reveal previously undetected differences between products of enzymatic and nonenzymatic base release, may shed light on the evolution and biological roles of AP endonucleases and AP lyases.
Keywords: AP endonucleases; AP lyases; N7-methylguanine; abasic sites; base excision repair.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. - PubMed
-
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. - PubMed
-
- Talpaert-Borlé M. Formation, detection and repair of AP sites. Mutat Res. 1987;181:45–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

