Differences in adaptive dynamics determine the success of virus variants that propagate together
- PMID: 29340211
- PMCID: PMC5761584
- DOI: 10.1093/ve/vex043
Differences in adaptive dynamics determine the success of virus variants that propagate together
Abstract
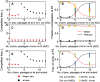
Virus fitness is a complex parameter that results from the interaction of virus-specific characters (e.g. intracellular growth rate, adsorption rate, virion extracellular stability, and tolerance to mutations) with others that depend on the underlying fitness landscape and the internal structure of the whole population. Individual mutants usually have lower fitness values than the complex population from which they come from. When they are propagated and allowed to attain large population sizes for a sufficiently long time, they approach mutation-selection equilibrium with the concomitant fitness gains. The optimization process follows dynamics that vary among viruses, likely due to differences in any of the parameters that determine fitness values. As a consequence, when different mutants spread together, the number of generations experienced by each of them prior to co-propagation may determine its particular fate. In this work we attempt a clarification of the effect of different levels of population diversity in the outcome of competition dynamics. To this end, we analyze the behavior of two mutants of the RNA bacteriophage Qβ that co-propagate with the wild-type virus. When both competitor viruses are clonal, the mutants rapidly outcompete the wild type. However, the outcome in competitions performed with partially optimized virus populations depends on the distance of the competitors to their clonal origin. We also implement a theoretical population dynamics model that describes the evolution of a heterogeneous population of individuals, each characterized by a fitness value, subjected to subsequent cycles of replication and mutation. The experimental results are explained in the framework of our theoretical model under two non-excluding, likely complementary assumptions: (1) The relative advantage of both competitors changes as populations approach mutation-selection equilibrium, as a consequence of differences in their growth rates and (2) one of the competitors is more robust to mutations than the other. The main conclusion is that the nearness of an RNA virus population to mutation-selection equilibrium is a key factor determining the fate of particular mutants arising during replication.
Keywords: computational models; error rate; fitness; population bottlenecks; population dynamics; virus adaptation.
Figures




Similar articles
-
Intra-Population Competition during Adaptation to Increased Temperature in an RNA Bacteriophage.Int J Mol Sci. 2021 Jun 24;22(13):6815. doi: 10.3390/ijms22136815. Int J Mol Sci. 2021. PMID: 34202838 Free PMC article.
-
Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures.Pathogens. 2019 Jun 18;8(2):80. doi: 10.3390/pathogens8020080. Pathogens. 2019. PMID: 31216651 Free PMC article.
-
Phenotypic effect of mutations in evolving populations of RNA molecules.BMC Evol Biol. 2010 Feb 17;10:46. doi: 10.1186/1471-2148-10-46. BMC Evol Biol. 2010. PMID: 20163698 Free PMC article.
-
Rational evolutionary design: the theory of in vitro protein evolution.Adv Protein Chem. 2000;55:79-160. doi: 10.1016/s0065-3233(01)55003-2. Adv Protein Chem. 2000. PMID: 11050933 Review.
-
Evolution of foot-and-mouth disease virus.Virus Res. 2003 Jan;91(1):47-63. doi: 10.1016/s0168-1702(02)00259-9. Virus Res. 2003. PMID: 12527437 Review.
Cited by
-
Cytosine drives evolution of SARS-CoV-2.Environ Microbiol. 2020 Jun;22(6):1977-1985. doi: 10.1111/1462-2920.15025. Epub 2020 Apr 27. Environ Microbiol. 2020. PMID: 32291894 Free PMC article. No abstract available.
-
The balance between fitness advantages and costs drives adaptation of bacteriophage Qβ to changes in host density at different temperatures.Front Microbiol. 2023 May 25;14:1197085. doi: 10.3389/fmicb.2023.1197085. eCollection 2023. Front Microbiol. 2023. PMID: 37303783 Free PMC article.
-
Standing Genetic Diversity and Transmission Bottleneck Size Drive Adaptation in Bacteriophage Qβ.Int J Mol Sci. 2022 Aug 9;23(16):8876. doi: 10.3390/ijms23168876. Int J Mol Sci. 2022. PMID: 36012143 Free PMC article.
-
Evolutionary Adaptation of an RNA Bacteriophage to Repeated Freezing and Thawing Cycles.Int J Mol Sci. 2024 Apr 29;25(9):4863. doi: 10.3390/ijms25094863. Int J Mol Sci. 2024. PMID: 38732084 Free PMC article.
-
Intra-Population Competition during Adaptation to Increased Temperature in an RNA Bacteriophage.Int J Mol Sci. 2021 Jun 24;22(13):6815. doi: 10.3390/ijms22136815. Int J Mol Sci. 2021. PMID: 34202838 Free PMC article.
References
-
- Aguirre J., Manrubia S. C. (2007) ‘Out-of-Equilibrium Competitive Dynamics of Quasispecies’, Europhysics Letters (Epl) , 77: 38001.
-
- Aguirre J., Lázaro E., Manrubia S. C. (2009) ‘A Trade-off between Fast Growth and Diversity Generation Limits the Optimization of Viral Quasispecies’, Journal of Theoretical Biology, 261: 148–55. - PubMed
-
- Arribas M., Cabanillas L., Lázaro E. (2011) ‘Identification of Mutations Conferring 5-Azacytidine Resistance in Bacteriophage Qβ’, Virology, 417: 343–52. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

