An automated framework for QSAR model building
- PMID: 29340790
- PMCID: PMC5770354
- DOI: 10.1186/s13321-017-0256-5
An automated framework for QSAR model building
Abstract
Background: In-silico quantitative structure-activity relationship (QSAR) models based tools are widely used to screen huge databases of compounds in order to determine the biological properties of chemical molecules based on their chemical structure. With the passage of time, the exponentially growing amount of synthesized and known chemicals data demands computationally efficient automated QSAR modeling tools, available to researchers that may lack extensive knowledge of machine learning modeling. Thus, a fully automated and advanced modeling platform can be an important addition to the QSAR community.
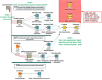
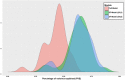
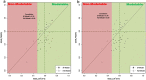
Results: In the presented workflow the process from data preparation to model building and validation has been completely automated. The most critical modeling tasks (data curation, data set characteristics evaluation, variable selection and validation) that largely influence the performance of QSAR models were focused. It is also included the ability to quickly evaluate the feasibility of a given data set to be modeled. The developed framework is tested on data sets of thirty different problems. The best-optimized feature selection methodology in the developed workflow is able to remove 62-99% of all redundant data. On average, about 19% of the prediction error was reduced by using feature selection producing an increase of 49% in the percentage of variance explained (PVE) compared to models without feature selection. Selecting only the models with a modelability score above 0.6, average PVE scores were 0.71. A strong correlation was verified between the modelability scores and the PVE of the models produced with variable selection.
Conclusions: We developed an extendable and highly customizable fully automated QSAR modeling framework. This designed workflow does not require any advanced parameterization nor depends on users decisions or expertise in machine learning/programming. With just a given target or problem, the workflow follows an unbiased standard protocol to develop reliable QSAR models by directly accessing online manually curated databases or by using private data sets. The other distinctive features of the workflow include prior estimation of data modelability to avoid time-consuming modeling trials for non modelable data sets, an efficient variable selection procedure and the facility of output availability at each modeling task for the diverse application and reproduction of historical predictions. The results reached on a selection of thirty QSAR problems suggest that the approach is capable of building reliable models even for challenging problems.
Keywords: Data set modelability; Feature selection; KNIME; Machine learning; Quantitative structure–activity relationship (QSAR); Random forests; Support vector machines; Variable importance.
Figures





References
-
- Matsumoto A, Aoki S, Ohwada H. Comparison of random forest and SVM for raw data in drug discovery: prediction of radiation protection and toxicity case study. Int J Mach Learn Comput. 2016;6(2):145–148. doi: 10.18178/ijmlc.2016.6.2.589. - DOI
-
- Mantus E. Toxicity testing in the 21st century. Alttox Org. 2007
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

