Trans-acting translational regulatory RNA binding proteins
- PMID: 29341429
- PMCID: PMC5947564
- DOI: 10.1002/wrna.1465
Trans-acting translational regulatory RNA binding proteins
Abstract
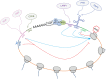
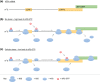
The canonical molecular machinery required for global mRNA translation and its control has been well defined, with distinct sets of proteins involved in the processes of translation initiation, elongation and termination. Additionally, noncanonical, trans-acting regulatory RNA-binding proteins (RBPs) are necessary to provide mRNA-specific translation, and these interact with 5' and 3' untranslated regions and coding regions of mRNA to regulate ribosome recruitment and transit. Recently it has also been demonstrated that trans-acting ribosomal proteins direct the translation of specific mRNAs. Importantly, it has been shown that subsets of RBPs often work in concert, forming distinct regulatory complexes upon different cellular perturbation, creating an RBP combinatorial code, which through the translation of specific subsets of mRNAs, dictate cell fate. With the development of new methodologies, a plethora of novel RNA binding proteins have recently been identified, although the function of many of these proteins within mRNA translation is unknown. In this review we will discuss these methodologies and their shortcomings when applied to the study of translation, which need to be addressed to enable a better understanding of trans-acting translational regulatory proteins. Moreover, we discuss the protein domains that are responsible for RNA binding as well as the RNA motifs to which they bind, and the role of trans-acting ribosomal proteins in directing the translation of specific mRNAs. This article is categorized under: RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes Translation > Translation Regulation Translation > Translation Mechanisms.
Keywords: IRES uORF; RNA binding protein; interactome capture; mRNA translation; protein synthesis; terminal oligopyrimidine tract.
© 2018 Medical Research Council and University of Cambridge. WIREs RNA published by Wiley Periodicals, Inc.
Figures

References
-
- Anger, A. M. , Armache, J.‐P. , Berninghausen, O. , Habeck, M. , Subklewe, M. , Wilson, D. N. , & Beckmann, R. (2013). Structures of the human and drosophila 80S ribosome. Nature, 497, 80–85. - PubMed
-
- Aoki, K. , Adachi, S. , Homoto, M. , Kusano, H. , Koike, K. , & Natsume, T. (2013). LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Letters, 587, 2173–2178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

