Expression of the lux genes in Streptococcus pneumoniae modulates pilus expression and virulence
- PMID: 29342160
- PMCID: PMC5771582
- DOI: 10.1371/journal.pone.0189426
Expression of the lux genes in Streptococcus pneumoniae modulates pilus expression and virulence
Abstract
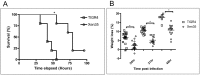
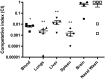
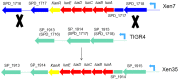
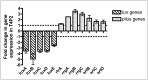
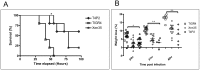
Bioluminescence has been harnessed for use in bacterial reporter systems and for in vivo imaging of infection in animal models. Strain Xen35, a bioluminescent derivative of Streptococcus pneumoniae serotype 4 strain TIGR4 was previously constructed for use for in vivo imaging of infections in animal models. We have shown that strain Xen35 is less virulent than its parent TIGR4 and that this is associated with the expression of the genes for bioluminescence. The expression of the luxA-E genes in the pneumococcus reduces virulence and down regulates the expression of the pneumococcal pilus.
Conflict of interest statement
Figures



References
-
- van der Linden M, Al-Lahham A, Nicklas W, Reinert RR. Molecular characterization of pneumococcal isolates from pets and laboratory animals. PLoS One. 2009;4(12):e8286 doi: 10.1371/journal.pone.0008286 - DOI - PMC - PubMed
-
- Chi F, Leider M, Leendertz F, Bergmann C, Boesch C, Schenk S, et al. New Streptococcus pneumoniae clones in deceased wild chimpanzees. J Bacteriol. 2007;189(16):6085–8. doi: 10.1128/JB.00468-07 - DOI - PMC - PubMed
-
- Havarstein LS, Gaustad P, Nes IF, Morrison DA. Identification of the streptococcal competence- pheromone receptor. Molecular Microbiology. 1996;21(4):863–9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

