Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions
- PMID: 29342876
- PMCID: PMC5796189
- DOI: 10.3390/ijms19010241
Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions
Abstract
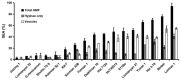
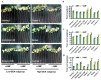
Arbuscular mycorrhizal fungi (AMF) play an important role in nutrient cycling processes and plant stress resistance. To evaluate the effect of Rhizophagus irregularis CD1 on plant growth promotion (PGP) and Verticillium wilt disease, the symbiotic efficiency of AMF (SEA) was first investigated over a range of 3% to 94% in 17 cotton varieties. The high-SEA subgroup had significant PGP effects in a greenhouse. From these results, the highest-SEA variety of Lumian 1 was selected for a two-year field assay. Consistent with the performance from the greenhouse, the AMF-mediated PGP of Lumian 1 also produced significant results, including an increased plant height, stem diameter, number of petioles, and phosphorus content. Compared with the mock treatment, AMF colonization obviously inhibited the symptom development of Verticillium dahliae and more strongly elevated the expression of pathogenesis-related genes and lignin synthesis-related genes. These results suggest that AMF colonization could lead to the mycorrhiza-induced resistance (MIR) of Lumian 1 to V. dahliae. Interestingly, our results indicated that the AMF endosymbiont could directly inhibit the growth of phytopathogenic fungi including V. dahliae by releasing undefined volatiles. In summary, our results suggest that stronger effects of AMF application result from the high-SEA.
Keywords: Gossypium hirsutum; Verticillium wilt; antifungal activity; mycorrhizal colonization; plant growth promotion; resistance; symbiotic efficiency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

