Implementing genome-driven personalized cardiology in clinical practice
- PMID: 29343412
- PMCID: PMC5820118
- DOI: 10.1016/j.yjmcc.2018.01.008
Implementing genome-driven personalized cardiology in clinical practice
Abstract
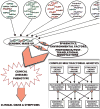
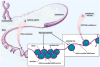
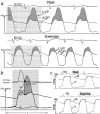
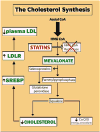
Genomics designates the coordinated investigation of a large number of genes in the context of a biological process or disease. It may be long before we attain comprehensive understanding of the genomics of common complex cardiovascular diseases (CVDs) such as inherited cardiomyopathies, valvular diseases, primary arrhythmogenic conditions, congenital heart syndromes, hypercholesterolemia and atherosclerotic heart disease, hypertensive syndromes, and heart failure with preserved/reduced ejection fraction. Nonetheless, as genomics is evolving rapidly, it is constructive to survey now pertinent concepts and breakthroughs. Today, clinical multimodal electronic medical/health records (EMRs/EHRs) incorporating genomic information establish a continuously-learning, vast knowledge-network with seamless cycling between clinical application and research. It can inform insights into specific pathogenetic pathways, guide biomarker-assisted precise diagnoses and individualized treatments, and stratify prognoses. Complex CVDs blend multiple interacting genomic variants, epigenetics, and environmental risk-factors, engendering progressions of multifaceted disease-manifestations, including clinical symptoms and signs. There is no straight-line linkage between genetic cause(s) or causal gene-variant(s) and disease phenotype(s). Because of interactions involving modifier-gene influences, (micro)-environmental, and epigenetic effects, the same variant may actually produce dissimilar abnormalities in different individuals. Implementing genome-driven personalized cardiology in clinical practice reveals that the study of CVDs at the level of molecules and cells can yield crucial clinical benefits. Complementing evidence-based medicine guidelines from large ("one-size fits all") randomized controlled trials, genomics-based personalized or precision cardiology is a most-creditable paradigm: It provides customizable approaches to prevent, diagnose, and manage CVDs with treatments directly/precisely aimed at causal defects identified by high-throughput genomic technologies. They encompass stem cell and gene therapies exploiting CRISPR-Cas9-gene-editing, and metabolomic-pharmacogenomic therapeutic modalities, precisely fine-tuned for the individual patient. Following the Human Genome Project, many expected genomics technology to provide imminent solutions to intractable medical problems, including CVDs. This eagerness has reaped some disappointment that advances have not yet materialized to the degree anticipated. Undoubtedly, personalized genetic/genomics testing is an emergent technology that should not be applied without supplementary phenotypic/clinical information: Genotype≠Phenotype. However, forthcoming advances in genomics will naturally build on prior attainments and, combined with insights into relevant epigenetics and environmental factors, can plausibly eradicate intractable CVDs, improving human health and well-being.
Keywords: Biomarkers; Genome sequencing; Genome-& phenome-wide association studies (GWAS/PheWAS); Genomic decoding of phenotypic diversity; Hemodynamics & myocardial mechanics phenome; Personalized or precision medicine.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Genomic translational research: Paving the way to individualized cardiac functional analyses and personalized cardiology.Int J Cardiol. 2017 Mar 1;230:384-401. doi: 10.1016/j.ijcard.2016.12.097. Epub 2016 Dec 21. Int J Cardiol. 2017. PMID: 28057368 Free PMC article. Review.
-
Precision Medicine in Pediatric Cardiology.Pediatr Ann. 2022 Oct;51(10):e390-e395. doi: 10.3928/19382359-20220803-05. Epub 2022 Oct 1. Pediatr Ann. 2022. PMID: 36215086 Review.
-
Big Data and Genome Editing Technology: A New Paradigm of Cardiovascular Genomics.Curr Cardiol Rev. 2017;13(4):301-304. doi: 10.2174/1573403X13666170804152432. Curr Cardiol Rev. 2017. PMID: 28782493 Free PMC article. Review.
-
Cardiovascular pharmacogenetics: a promise for genomically-guided therapy and personalized medicine.Clin Genet. 2017 Mar;91(3):355-370. doi: 10.1111/cge.12881. Epub 2016 Nov 30. Clin Genet. 2017. PMID: 27714756 Review.
-
Genomic insights into heart health: Exploring the genetic basis of cardiovascular disease.Curr Probl Cardiol. 2024 Jan;49(1 Pt C):102182. doi: 10.1016/j.cpcardiol.2023.102182. Epub 2023 Oct 30. Curr Probl Cardiol. 2024. PMID: 37913933 Review.
Cited by
-
The new era of whole-exome sequencing in congenital heart disease: brand-new insights into rare pathogenic variants.J Thorac Dis. 2018 Jun;10(Suppl 17):S1923-S1929. doi: 10.21037/jtd.2018.05.56. J Thorac Dis. 2018. PMID: 30023082 Free PMC article. No abstract available.
-
Clinical-pathological correlations of BAV and the attendant thoracic aortopathies. Part 2: Pluridisciplinary perspective on their genetic and molecular origins.J Mol Cell Cardiol. 2019 Aug;133:233-246. doi: 10.1016/j.yjmcc.2019.05.022. Epub 2019 Jun 6. J Mol Cell Cardiol. 2019. PMID: 31175858 Free PMC article. Review.
-
The Current Therapeutic Role of Chromatin Remodeling for the Prognosis and Treatment of Heart Failure.Biomedicines. 2023 Feb 16;11(2):579. doi: 10.3390/biomedicines11020579. Biomedicines. 2023. PMID: 36831115 Free PMC article. Review.
-
Interplay Between Gut Microbiota and Amino Acid Metabolism in Heart Failure.Front Cardiovasc Med. 2021 Oct 21;8:752241. doi: 10.3389/fcvm.2021.752241. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34746265 Free PMC article. Review.
-
Gene surgery: Potential applications for human diseases.EXCLI J. 2019 Oct 11;18:908-930. doi: 10.17179/excli2019-1833. eCollection 2019. EXCLI J. 2019. PMID: 31762718 Free PMC article. Review.
References
-
- Pasipoularides A. Heart's vortex: Intracardiac blood flow phenomena. Shelton, CT, USA: People'sMedical Publishing House; 2010.
-
- Huijgen R, Vissers MN, Defesche JC, et al. Familial hypercholesterolemia: current treatment and advances in management. Expert Rev Cardiovasc Ther. 2008;6:567–81. - PubMed
-
- Mark M, Rijli F, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

