Oxysterol Sensing through the Receptor GPR183 Promotes the Lymphoid-Tissue-Inducing Function of Innate Lymphoid Cells and Colonic Inflammation
- PMID: 29343433
- PMCID: PMC5772175
- DOI: 10.1016/j.immuni.2017.11.020
Oxysterol Sensing through the Receptor GPR183 Promotes the Lymphoid-Tissue-Inducing Function of Innate Lymphoid Cells and Colonic Inflammation
Abstract
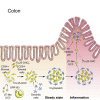
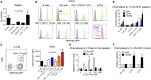
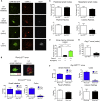
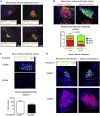
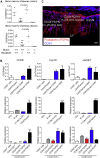
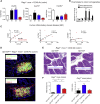
Group 3 innate lymphoid cells (ILC3s) sense environmental signals and are critical for tissue integrity in the intestine. Yet, which signals are sensed and what receptors control ILC3 function remain poorly understood. Here, we show that ILC3s with a lymphoid-tissue-inducer (LTi) phenotype expressed G-protein-coupled receptor 183 (GPR183) and migrated to its oxysterol ligand 7α,25-hydroxycholesterol (7α,25-OHC). In mice lacking Gpr183 or 7α,25-OHC, ILC3s failed to localize to cryptopatches (CPs) and isolated lymphoid follicles (ILFs). Gpr183 deficiency in ILC3s caused a defect in CP and ILF formation in the colon, but not in the small intestine. Localized oxysterol production by fibroblastic stromal cells provided an essential signal for colonic lymphoid tissue development, and inflammation-induced increased oxysterol production caused colitis through GPR183-mediated cell recruitment. Our findings show that GPR183 promotes lymphoid organ development and indicate that oxysterol-GPR183-dependent positioning within tissues controls ILC3 activity and intestinal homeostasis.
Keywords: EBI2; GPR183; cell migration; colon; inflammation; innate lymphoid cells; lymphoid tissue; oxysterols.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

