Identification of Poxvirus Genome Uncoating and DNA Replication Factors with Mutually Redundant Roles
- PMID: 29343579
- PMCID: PMC5972866
- DOI: 10.1128/JVI.02152-17
Identification of Poxvirus Genome Uncoating and DNA Replication Factors with Mutually Redundant Roles
Abstract
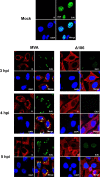
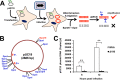
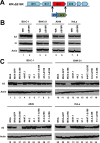
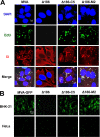
Genome uncoating is essential for replication of most viruses. For poxviruses, the process is divided into two stages: removal of the envelope, allowing early gene expression, and breaching of the core wall, allowing DNA release, replication, and late gene expression. Subsequent studies showed that the host proteasome and the viral D5 protein, which has an essential role in DNA replication, are required for vaccinia virus (VACV) genome uncoating. In a search for additional VACV uncoating proteins, we noted a report that described a defect in DNA replication and late expression when the gene encoding a 68-kDa ankyrin repeat/F-box protein (68k-ank), associated with the cellular SCF (Skp1, cullin1, F-box-containing complex) ubiquitin ligase complex, was deleted from the attenuated modified vaccinia virus Ankara (MVA). Here we showed that the 68k-ank deletion mutant exhibited diminished genome uncoating, formation of DNA prereplication sites, and degradation of viral cores as well as an additional, independent defect in DNA synthesis. Deletion of the 68k-ank homolog of VACV strain WR, however, was without effect, suggesting the existence of compensating genes. By inserting VACV genes into an MVA 68k-ank deletion mutant, we discovered that M2, a member of the poxvirus immune evasion (PIE) domain superfamily and a regulator of NF-κB, and C5, a member of the BTB/Kelch superfamily associated with cullin-3-based ligase complexes, independently rescued the 68k-ank deletion phenotype. Thus, poxvirus uncoating and DNA replication are intertwined processes involving at least three viral proteins with mutually redundant functions in addition to D5.IMPORTANCE Poxviruses comprise a family of large DNA viruses that infect vertebrates and invertebrates and cause diseases of medical and zoological importance. Poxviruses, unlike most other DNA viruses, replicate in the cytoplasm, and their large genomes usually encode 200 or more proteins with diverse functions. About 90 genes may be essential for chordopoxvirus replication based either on their conservation or individual gene deletion studies. However, this number may underestimate the true number of essential functions because of redundancy. Here we show that any one of three seemingly unrelated and individually nonessential proteins is required for the incompletely understood processes of genome uncoating and DNA replication, an example of synthetic lethality. Thus, poxviruses appear to have a complex genetic interaction network that has not been fully appreciated and which will require multifactor deletion screens to assess.
Keywords: gene redundancy; genome replication; genome uncoating; poxvirus replication; recombinant DNA technology; synthetic lethality; vaccinia virus.
Copyright © 2018 American Society for Microbiology.
Figures




References
-
- Joklik WK. 1964. The intracellular uncoating of poxvirus DNA. I. The fate of radioactively-labeled rabbitpox virus. J Mol Biol 8:263–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

