The protective role of DOT1L in UV-induced melanomagenesis
- PMID: 29343685
- PMCID: PMC5772495
- DOI: 10.1038/s41467-017-02687-7
The protective role of DOT1L in UV-induced melanomagenesis
Abstract
The DOT1L histone H3 lysine 79 (H3K79) methyltransferase plays an oncogenic role in MLL-rearranged leukemogenesis. Here, we demonstrate that, in contrast to MLL-rearranged leukemia, DOT1L plays a protective role in ultraviolet radiation (UVR)-induced melanoma development. Specifically, the DOT1L gene is located in a frequently deleted region and undergoes somatic mutation in human melanoma. Specific mutations functionally compromise DOT1L methyltransferase enzyme activity leading to reduced H3K79 methylation. Importantly, in the absence of DOT1L, UVR-induced DNA damage is inefficiently repaired, so that DOT1L loss promotes melanoma development in mice after exposure to UVR. Mechanistically, DOT1L facilitates DNA damage repair, with DOT1L-methylated H3K79 involvement in binding and recruiting XPC to the DNA damage site for nucleotide excision repair (NER). This study indicates that DOT1L plays a protective role in UVR-induced melanomagenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



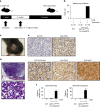
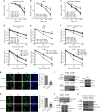
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

