Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding
- PMID: 29345219
- PMCID: PMC5857379
- DOI: 10.1099/mgen.0.000152
Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding
Abstract
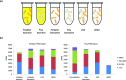
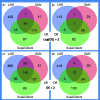
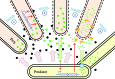
Predation is a fundamental ecological process, but within most microbial ecosystems the molecular mechanisms of predation remain poorly understood. We investigated transcriptome changes associated with the predation of Escherichia coli by the myxobacterium Myxococcus xanthus using mRNA sequencing. Exposure to pre-killed prey significantly altered expression of 1319 predator genes. However, the transcriptional response to living prey was minimal, with only 12 genes being significantly up-regulated. The genes most induced by prey presence (kdpA and kdpB, members of the kdp regulon) were confirmed by reverse transcriptase quantitative PCR to be regulated by osmotic shock in M. xanthus, suggesting indirect sensing of prey. However, the prey showed extensive transcriptome changes when co-cultured with predator, with 40 % of its genes (1534) showing significant changes in expression. Bacteriolytic M. xanthus culture supernatant and secreted outer membrane vesicles (OMVs) also induced changes in expression of large numbers of prey genes (598 and 461, respectively). Five metabolic pathways were significantly enriched in prey genes up-regulated on exposure to OMVs, supernatant and/or predatory cells, including those for ribosome and lipopolysaccharide production, suggesting that the prey cell wall and protein production are primary targets of the predator's attack. Our data suggest a model of the myxobacterial predatome (genes and proteins associated with predation) in which the predator constitutively produces secretions which disable its prey whilst simultaneously generating a signal that prey is present. That signal then triggers a regulated feeding response in the predator.
Keywords: Myxobacteria; antimicrobial activity; mixed culture; outer membrane vesicles; predatome; transcriptome.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Dynamics of Solitary Predation by Myxococcus xanthus on Escherichia coli Observed at the Single-Cell Level.Appl Environ Microbiol. 2020 Jan 21;86(3):e02286-19. doi: 10.1128/AEM.02286-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31704687 Free PMC article.
-
The Genetics of Prey Susceptibility to Myxobacterial Predation: A Review, Including an Investigation into Pseudomonas aeruginosa Mutations Affecting Predation by Myxococcus xanthus.Microb Physiol. 2021;31(2):57-66. doi: 10.1159/000515546. Epub 2021 Apr 1. Microb Physiol. 2021. PMID: 33794538 Review.
-
Interspecies conflict affects RNA expression.FEMS Microbiol Lett. 2018 May 1;365(10). doi: 10.1093/femsle/fny096. FEMS Microbiol Lett. 2018. PMID: 29648585
-
Myxobacterial vesicles death at a distance?Adv Appl Microbiol. 2011;75:1-31. doi: 10.1016/B978-0-12-387046-9.00001-3. Adv Appl Microbiol. 2011. PMID: 21807244 Review.
-
Predatory Strategies of Myxococcus xanthus: Prey Susceptibility to OMVs and Moonlighting Enzymes.Microorganisms. 2023 Mar 29;11(4):874. doi: 10.3390/microorganisms11040874. Microorganisms. 2023. PMID: 37110297 Free PMC article.
Cited by
-
The antibiotic crisis: How bacterial predators can help.Comput Struct Biotechnol J. 2020 Sep 15;18:2547-2555. doi: 10.1016/j.csbj.2020.09.010. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33033577 Free PMC article. Review.
-
Genome Analysis, Metabolic Potential, and Predatory Capabilities of Herpetosiphon llansteffanense sp. nov.Appl Environ Microbiol. 2018 Oct 30;84(22):e01040-18. doi: 10.1128/AEM.01040-18. Print 2018 Nov 15. Appl Environ Microbiol. 2018. PMID: 30194103 Free PMC article.
-
Development versus predation: Transcriptomic changes during the lifecycle of Myxococcus xanthus.Front Microbiol. 2022 Sep 26;13:1004476. doi: 10.3389/fmicb.2022.1004476. eCollection 2022. Front Microbiol. 2022. PMID: 36225384 Free PMC article.
-
Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio.Nat Commun. 2019 Sep 9;10(1):4086. doi: 10.1038/s41467-019-12051-6. Nat Commun. 2019. PMID: 31501441 Free PMC article.
-
Differential response to prey quorum signals indicates predatory specialization of myxobacteria and ability to predate Pseudomonas aeruginosa.Environ Microbiol. 2022 Mar;24(3):1263-1278. doi: 10.1111/1462-2920.15812. Epub 2021 Oct 21. Environ Microbiol. 2022. PMID: 34674390 Free PMC article.
References
-
- Whitworth DE. Myxobacteria: Multicellularity and Differentiation. Washington, DC: American Society for Microbiology; 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

