Efficient Fusion at Neutral pH by Human Immunodeficiency Virus gp41 Trimers Containing the Fusion Peptide and Transmembrane Domains
- PMID: 29345922
- PMCID: PMC6151270
- DOI: 10.1021/acs.biochem.7b00753
Efficient Fusion at Neutral pH by Human Immunodeficiency Virus gp41 Trimers Containing the Fusion Peptide and Transmembrane Domains
Abstract
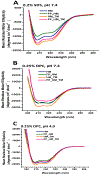
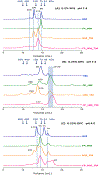
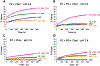
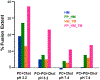
Human immunodeficiency virus (HIV) is membrane-enveloped, and an initial infection step is joining/fusion of viral and cell membranes. This step is catalyzed by gp41, which is a single-pass integral viral membrane protein. The protein contains an ∼170-residue ectodomain located outside the virus that is important for fusion and includes the fusion peptide (FP), N-helix, loop, C-helix, and viral membrane-proximal external region (MPER). The virion initially has noncovalent complexes between three gp41 ectodomains and three gp120 proteins. A gp120 contains ∼500 residues and functions to identify target T-cells and macrophages via binding to specific protein receptors of the target cell membrane. gp120 moves away from the gp41 ectodomain, and the ectodomain is thought to bind to the target cell membrane and mediate membrane fusion. The secondary and tertiary structures of the ectodomain are different in the initial complex with gp120 and the final state without gp120. There is not yet imaging of gp41 during fusion, so the temporal relationship between the gp41 and membrane structures is not known. This study describes biophysical and functional characterization of large gp41 constructs that include the ectodomain and transmembrane domain (TM). Significant fusion is observed of both neutral and anionic vesicles at neutral pH, which reflects the expected conditions of HIV/cell fusion. Fusion is enhanced by the FP, which in HIV/cell fusion likely contacts the host membrane, and the MPER and TM, which respectively interfacially contact and traverse the HIV membrane. Initial contact with vesicles is made by protein trimers that are in a native oligomeric state that reflects the initial complex with gp120 and also is commonly observed for the ectodomain without gp120. Circular dichroism data support helical structure for the N-helix, C-helix, and MPER and nonhelical structure for the FP and loop. Distributions of monomer, trimer, and hexamer states are observed by size-exclusion chromatography (SEC), with dependences on solubilizing detergent and construct. These SEC and other data are integrated into a refined working model of HIV/cell fusion that includes dissociation of the ectodomain into gp41 monomers followed by folding into hairpins that appose the two membranes, and subsequent fusion catalysis by trimers and hexamers of hairpins. The monomer and oligomer gp41 states may therefore satisfy dual requirements for HIV entry of membrane apposition and fusion.
Figures






Similar articles
-
Folded monomers and hexamers of the ectodomain of the HIV gp41 membrane fusion protein: potential roles in fusion and synergy between the fusion peptide, hairpin, and membrane-proximal external region.Biochemistry. 2014 Nov 25;53(46):7184-98. doi: 10.1021/bi501159w. Epub 2014 Nov 14. Biochemistry. 2014. PMID: 25372604 Free PMC article.
-
Conditional trimerization and lytic activity of HIV-1 gp41 variants containing the membrane-associated segments.Biochemistry. 2015 Mar 3;54(8):1589-99. doi: 10.1021/bi501376f. Epub 2015 Feb 13. Biochemistry. 2015. PMID: 25658332 Free PMC article.
-
A large HIV gp41 construct with trimer-of-hairpins structure exhibits V2E mutation-dominant attenuation of vesicle fusion and helicity very similar to V2E attenuation of HIV fusion and infection and supports: (1) hairpin stabilization of membrane apposition with larger distance for V2E; and (2) V2E dominance by an antiparallel β sheet with interleaved fusion peptide strands from two gp41 trimers.Biophys Chem. 2023 Feb;293:106933. doi: 10.1016/j.bpc.2022.106933. Epub 2022 Nov 24. Biophys Chem. 2023. PMID: 36508984 Free PMC article.
-
Biochemistry and biophysics of HIV-1 gp41 - membrane interactions and implications for HIV-1 envelope protein mediated viral-cell fusion and fusion inhibitor design.Curr Top Med Chem. 2011 Dec;11(24):2959-84. doi: 10.2174/156802611798808497. Curr Top Med Chem. 2011. PMID: 22044229 Free PMC article. Review.
-
HIV-1 gp41: mediator of fusion and target for inhibition.AIDS Rev. 2003 Oct-Dec;5(4):214-21. AIDS Rev. 2003. PMID: 15012000 Review.
Cited by
-
Hydrogen-Deuterium Exchange Supports Independent Membrane-Interfacial Fusion Peptide and Transmembrane Domains in Subunit 2 of Influenza Virus Hemagglutinin Protein, a Structured and Aqueous-Protected Connection between the Fusion Peptide and Soluble Ectodomain, and the Importance of Membrane Apposition by the Trimer-of-Hairpins Structure.Biochemistry. 2019 May 14;58(19):2432-2446. doi: 10.1021/acs.biochem.8b01272. Epub 2019 May 1. Biochemistry. 2019. PMID: 31008587 Free PMC article.
-
Broadly neutralizing antibodies for HIV prevention: a comprehensive review and future perspectives.Clin Microbiol Rev. 2024 Jun 13;37(2):e0015222. doi: 10.1128/cmr.00152-22. Epub 2024 Apr 30. Clin Microbiol Rev. 2024. PMID: 38687039 Free PMC article. Review.
-
Rapid 2H NMR Transverse Relaxation of Perdeuterated Lipid Acyl Chains of Membrane with Bound Viral Fusion Peptide Supports Large-Amplitude Motions of These Chains That Can Catalyze Membrane Fusion.Biochemistry. 2021 Sep 7;60(35):2637-2651. doi: 10.1021/acs.biochem.1c00316. Epub 2021 Aug 26. Biochemistry. 2021. PMID: 34436856 Free PMC article.
-
2H nuclear magnetic resonance spectroscopy supports larger amplitude fast motion and interference with lipid chain ordering for membrane that contains β sheet human immunodeficiency virus gp41 fusion peptide or helical hairpin influenza virus hemagglutinin fusion peptide at fusogenic pH.Biochim Biophys Acta Biomembr. 2020 Oct 1;1862(10):183404. doi: 10.1016/j.bbamem.2020.183404. Epub 2020 Jun 23. Biochim Biophys Acta Biomembr. 2020. PMID: 32585207 Free PMC article.
-
Insights into the mechanism of membrane fusion induced by the plant defense element, plant-specific insert.J Biol Chem. 2020 Oct 23;295(43):14548-14562. doi: 10.1074/jbc.RA120.014311. Epub 2020 Jul 10. J Biol Chem. 2020. PMID: 32651232 Free PMC article.
References
-
- Grewe C, Beck A, and Gelderblom HR (1990) HIV: early virus‐cell interactions, J. AIDS 3, 965–974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

