The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer
- PMID: 29346117
- PMCID: PMC5873885
- DOI: 10.1172/JCI97325
The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer
Abstract
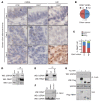
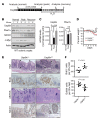
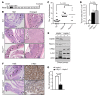
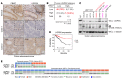
The tumor suppressor FBW7 targets oncoproteins such as c-MYC for ubiquitylation and is mutated in several human cancers. We noted that in a substantial percentage of colon cancers, FBW7 protein is undetectable despite the presence of FBW7 mRNA. To understand the molecular mechanism of FBW7 regulation in these cancers, we employed proteomics and identified the deubiquitinase (DUB) USP9X as an FBW7 interactor. USP9X antagonized FBW7 ubiquitylation, and Usp9x deletion caused Fbw7 destabilization. Mice lacking Usp9x in the gut showed reduced secretory cell differentiation and increased progenitor proliferation, phenocopying Fbw7 loss. In addition, Usp9x inactivation impaired intestinal regeneration and increased tumor burden in colitis-associated intestinal cancer. c-Myc heterozygosity abrogated increased progenitor proliferation and tumor burden in Usp9x-deficient mice, suggesting that Usp9x suppresses tumor formation by regulating Fbw7 protein stability and thereby reducing c-Myc. Thus, we identify a tumor suppressor mechanism in the mammalian intestine that arises from the posttranslational regulation of FBW7 by USP9X independent of somatic FBW7 mutations.
Keywords: Cell Biology; Colorectal cancer; Gastroenterology; Ubiquitin-proteosome system.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

