The Evolutionary History of Nebraska Deer Mice: Local Adaptation in the Face of Strong Gene Flow
- PMID: 29346646
- PMCID: PMC5905656
- DOI: 10.1093/molbev/msy004
The Evolutionary History of Nebraska Deer Mice: Local Adaptation in the Face of Strong Gene Flow
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
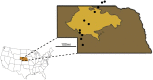
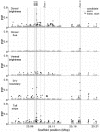
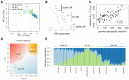
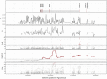
The interplay of gene flow, genetic drift, and local selective pressure is a dynamic process that has been well studied from a theoretical perspective over the last century. Wright and Haldane laid the foundation for expectations under an island-continent model, demonstrating that an island-specific beneficial allele may be maintained locally if the selection coefficient is larger than the rate of migration of the ancestral allele from the continent. Subsequent extensions of this model have provided considerably more insight. Yet, connecting theoretical results with empirical data has proven challenging, owing to a lack of information on the relationship between genotype, phenotype, and fitness. Here, we examine the demographic and selective history of deer mice in and around the Nebraska Sand Hills, a system in which variation at the Agouti locus affects cryptic coloration that in turn affects the survival of mice in their local habitat. We first genotyped 250 individuals from 11 sites along a transect spanning the Sand Hills at 660,000 single nucleotide polymorphisms across the genome. Using these genomic data, we found that deer mice first colonized the Sand Hills following the last glacial period. Subsequent high rates of gene flow have served to homogenize the majority of the genome between populations on and off the Sand Hills, with the exception of the Agouti pigmentation locus. Furthermore, mutations at this locus are strongly associated with the pigment traits that are strongly correlated with local soil coloration and thus responsible for cryptic coloration.
Figures


References
-
- Akaike H. 1974. New look at statistical-model identification. IEEE Trans Automat Contr. 196: 716–723. 10.1109/TAC.1974.1100705 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

