Bipartite Network Analysis of Gene Sharings in the Microbial World
- PMID: 29346651
- PMCID: PMC5888944
- DOI: 10.1093/molbev/msy001
Bipartite Network Analysis of Gene Sharings in the Microbial World
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
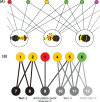
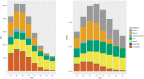
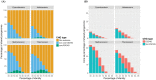
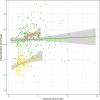
Extensive microbial gene flows affect how we understand virology, microbiology, medical sciences, genetic modification, and evolutionary biology. Phylogenies only provide a narrow view of these gene flows: plasmids and viruses, lacking core genes, cannot be attached to cellular life on phylogenetic trees. Yet viruses and plasmids have a major impact on cellular evolution, affecting both the gene content and the dynamics of microbial communities. Using bipartite graphs that connect up to 149,000 clusters of homologous genes with 8,217 related and unrelated genomes, we can in particular show patterns of gene sharing that do not map neatly with the organismal phylogeny. Homologous genes are recycled by lateral gene transfer, and multiple copies of homologous genes are carried by otherwise completely unrelated (and possibly nested) genomes, that is, viruses, plasmids and prokaryotes. When a homologous gene is present on at least one plasmid or virus and at least one chromosome, a process of "gene externalization," affected by a postprocessed selected functional bias, takes place, especially in Bacteria. Bipartite graphs give us a view of vertical and horizontal gene flow beyond classic taxonomy on a single very large, analytically tractable, graph that goes beyond the cellular Web of Life.
Figures
References
-
- Aleksandrzak-Piekarczyk T, Koryszewska-Bagińska A, Grynberg M, Nowak A, Cukrowska B, Kozakova H, Bardowski J.. 2016. Genomic and functional characterization of the unusual pLOCK 0919 plasmid harboring the spaCBA Pili cluster in Lactobacillus casei LOCK 0919. Genome Biol Evol. 81:202–217. - PMC - PubMed
-
- Alzahrani T, Horadam KJ.. 2016. Complex systems and networks In: Lü J, Yu X, Chen G, Yu W, editors. Understanding complex systems. Vol. 73 Berlin, Heidelberg: Springer Berlin Heidelberg.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

