Detecting Polygenic Adaptation in Admixture Graphs
- PMID: 29348143
- PMCID: PMC5887149
- DOI: 10.1534/genetics.117.300489
Detecting Polygenic Adaptation in Admixture Graphs
Abstract
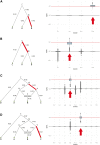
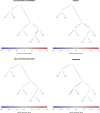
An open question in human evolution is the importance of polygenic adaptation: adaptive changes in the mean of a multifactorial trait due to shifts in allele frequencies across many loci. In recent years, several methods have been developed to detect polygenic adaptation using loci identified in genome-wide association studies (GWAS). Though powerful, these methods suffer from limited interpretability: they can detect which sets of populations have evidence for polygenic adaptation, but are unable to reveal where in the history of multiple populations these processes occurred. To address this, we created a method to detect polygenic adaptation in an admixture graph, which is a representation of the historical divergences and admixture events relating different populations through time. We developed a Markov chain Monte Carlo (MCMC) algorithm to infer branch-specific parameters reflecting the strength of selection in each branch of a graph. Additionally, we developed a set of summary statistics that are fast to compute and can indicate which branches are most likely to have experienced polygenic adaptation. We show via simulations that this method-which we call PolyGraph-has good power to detect polygenic adaptation, and applied it to human population genomic data from around the world. We also provide evidence that variants associated with several traits, including height, educational attainment, and self-reported unibrow, have been influenced by polygenic adaptation in different populations during human evolution.
Keywords: GWAS; admixture; complex traits; polygenic adaptation; selection.
Copyright © 2018 by the Genetics Society of America.
Figures






References
-
- Berg J. J., Zhang X., Coop G., 2017. Polygenic adaptation has impacted multiple anthropometric traits. bioRxiv. https://doi.org/10.1101/167551. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

