The α-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling
- PMID: 29348172
- PMCID: PMC5836114
- DOI: 10.1074/jbc.RA117.001516
The α-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling
Abstract
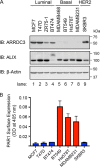
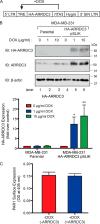
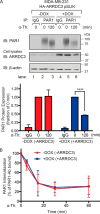
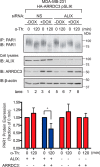
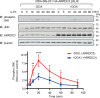
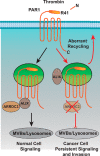
Aberrant G protein-coupled receptor (GPCR) expression and activation has been linked to tumor initiation, progression, invasion, and metastasis. However, compared with other cancer drivers, the exploitation of GPCRs as potential therapeutic targets has been largely ignored, despite the fact that GPCRs are highly druggable. Therefore, to advance the potential status of GPCRs as therapeutic targets, it is important to understand how GPCRs function together with other cancer drivers during tumor progression. We now report that the α-arrestin domain-containing protein-3 (ARRDC3) acts as a tumor suppressor in part by controlling signaling and trafficking of the GPCR, protease-activated receptor-1 (PAR1). In a series of highly invasive basal-like breast carcinomas, we found that expression of ARRDC3 is suppressed whereas PAR1 is aberrantly overexpressed because of defective lysosomal sorting that results in persistent signaling. Using a lentiviral doxycycline-inducible system, we demonstrate that re-expression of ARRDC3 in invasive breast carcinoma is sufficient to restore normal PAR1 trafficking through the ALG-interacting protein X (ALIX)-dependent lysosomal degradative pathway. We also show that ARRDC3 re-expression attenuates PAR1-stimulated persistent signaling of c-Jun N-terminal kinase (JNK) in invasive breast cancer. Remarkably, restoration of ARRDC3 expression significantly reduced activated PAR1-induced breast carcinoma invasion, which was also dependent on JNK signaling. These findings are the first to identify a critical link between the tumor suppressor ARRDC3 and regulation of GPCR trafficking and signaling in breast cancer.
Keywords: GPCR; Rho (Rho GTPase); beta-arrestin; breast cancer; c-Jun N-terminal kinase (JNK); protease; protease-activated receptor; thrombin; trafficking.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Prickett T. D., Wei X., Cardenas-Navia I., Teer J. K., Lin J. C., Walia V., Gartner J., Jiang J., Cherukuri P. F., Molinolo A., Davies M. A., Gershenwald J. E., Stemke-Hale K., Rosenberg S. A., Margulies E. H., and Samuels Y. (2011) Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat. Genet. 43, 1119–1126 10.1038/ng.950 - DOI - PMC - PubMed
-
- Kan Z., Jaiswal B. S., Stinson J., Janakiraman V., Bhatt D., Stern H. M., Yue P., Haverty P. M., Bourgon R., Zheng J., Moorhead M., Chaudhuri S., Tomsho L. P., Peters B. A., Pujara K., et al. (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 10.1038/nature09208 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

