Development of a predictive miRNA signature for breast cancer risk among high-risk women
- PMID: 29348816
- PMCID: PMC5762501
- DOI: 10.18632/oncotarget.22750
Development of a predictive miRNA signature for breast cancer risk among high-risk women
Abstract
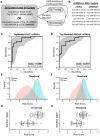
Significant limitations exist in our ability to predict breast cancer risk at the individual level. Circulating microRNAs (C-miRNAs) have emerged as measurable biomarkers (liquid biopsies) for cancer detection. We evaluated the ability of C-miRNAs to identify women most likely to develop breast cancer by profiling miRNA from serum obtained long before diagnosis. 24 breast cancer cases and controls (matched for risk and age) were identified from women enrolled in the High-Risk Breast Program at the UVM Cancer Center. Isolated RNA from serum was profiled for over 2500 human miRNAs. The miRNA expression data were input into a stepwise linear regression model to discover a multivariable miRNA signature that predicts long-term risk of breast cancer. 25 candidate miRNAs were identified that individually classified cases and controls based on statistical methodologies. A refined 6-miRNA risk-signature was discovered following regression modeling that distinguishes cases and controls (AUC0.896, CI 0.804-0.988) in this cohort. A functional relationship between miRNAs that cluster together when cases are contrasted against controls was suggested and confirmed by pathway analyses. The discovered 6 miRNA risk-signature can discriminate high-risk women who ultimately develop breast cancer from those who remain cancer-free, improving current risk assessment models. Future studies will focus on functional analysis of the miRNAs in this signature and testing in larger cohorts. We propose that the combined signature is highly significant for predicting cancer risk, and worthy of further screening in larger, independent clinical cohorts.
Keywords: benign breast disease; high risk breast cancer; liquid biopsy; microRNA; risk signature.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no potential conflicts of interest.
Figures



References
-
- Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–86. - PubMed
-
- Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction. Cancer. 1994;73:643–51. - PubMed
-
- Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23:1111–30. - PubMed
-
- Costantino JP, Gail MH, Pee D, Anderson S, Redmond CK, Benichou J, Wieand HS. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst. 1999;91:1541–8. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

