Iterative random forests to discover predictive and stable high-order interactions
- PMID: 29351989
- PMCID: PMC5828575
- DOI: 10.1073/pnas.1711236115
Iterative random forests to discover predictive and stable high-order interactions
Abstract
Genomics has revolutionized biology, enabling the interrogation of whole transcriptomes, genome-wide binding sites for proteins, and many other molecular processes. However, individual genomic assays measure elements that interact in vivo as components of larger molecular machines. Understanding how these high-order interactions drive gene expression presents a substantial statistical challenge. Building on random forests (RFs) and random intersection trees (RITs) and through extensive, biologically inspired simulations, we developed the iterative random forest algorithm (iRF). iRF trains a feature-weighted ensemble of decision trees to detect stable, high-order interactions with the same order of computational cost as the RF. We demonstrate the utility of iRF for high-order interaction discovery in two prediction problems: enhancer activity in the early Drosophila embryo and alternative splicing of primary transcripts in human-derived cell lines. In Drosophila, among the 20 pairwise transcription factor interactions iRF identifies as stable (returned in more than half of bootstrap replicates), 80% have been previously reported as physical interactions. Moreover, third-order interactions, e.g., between Zelda (Zld), Giant (Gt), and Twist (Twi), suggest high-order relationships that are candidates for follow-up experiments. In human-derived cells, iRF rediscovered a central role of H3K36me3 in chromatin-mediated splicing regulation and identified interesting fifth- and sixth-order interactions, indicative of multivalent nucleosomes with specific roles in splicing regulation. By decoupling the order of interactions from the computational cost of identification, iRF opens additional avenues of inquiry into the molecular mechanisms underlying genome biology.
Keywords: genomics; high-order interaction; interpretable machine learning; random forests; stability.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
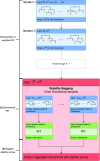

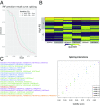
Comment in
-
Classification and interaction in random forests.Proc Natl Acad Sci U S A. 2018 Feb 20;115(8):1690-1692. doi: 10.1073/pnas.1800256115. Epub 2018 Feb 12. Proc Natl Acad Sci U S A. 2018. PMID: 29440440 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

