Subtype-Specific Selection for Resistance to Fluoroquinolones but Not to Tetracyclines Is Evident in Campylobacter jejuni Isolates from Beef Cattle in Confined Feeding Operations in Southern Alberta, Canada
- PMID: 29352087
- PMCID: PMC5861835
- DOI: 10.1128/AEM.02713-17
Subtype-Specific Selection for Resistance to Fluoroquinolones but Not to Tetracyclines Is Evident in Campylobacter jejuni Isolates from Beef Cattle in Confined Feeding Operations in Southern Alberta, Canada
Abstract
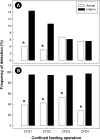
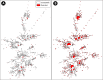
Campylobacter jejuni was longitudinally isolated from beef cattle housed in four confined feeding operations (CFOs) in Southern Alberta, Canada, over 18 months. All of the cattle were administered a variety of antimicrobial agents (AMAs) nontherapeutically and metaphylactically during their time in the CFOs. In total, 7,966 C. jejuni isolates were recovered from cattle. More animals were colonized by the bacterium after >60 days in the CFO (interim) than were individuals upon entry at the CFO (arrival). Subtyping and resistance to seven AMAs were determined for 1,832 (23.0%) and 1,648 (20.7%) isolates, respectively. Increases in the proportion of isolates resistant to tetracycline were observed at all four CFOs between sample times and to ciprofloxacin and nalidixic acid at one or more CFOs. The vast majority of isolates resistant to tetracycline carried tetO, whereas ciprofloxacin resistance was predominantly attributed to mutations in the gyrA gene. Although considerable diversity was observed, a majority of C. jejuni isolates belonged to one of five predominant subtype clusters. There was no difference in subtype diversity by CFO, but the population structure differed between sample times. Selection for resistance to ciprofloxacin and nalidixic acid was subtype dependent, whereas selection for resistance to tetracycline was not. The findings indicate that a proportion of cattle entering CFOs carry resistant C. jejuni subtypes, and the characteristics of beef cattle CFOs facilitate transmission/proliferation of diverse subtypes, including those resistant to AMAs, which coupled with the densities of CFOs likely contribute to the high rates of cattle-associated campylobacteriosis in Southern Alberta.IMPORTANCE A small proportion of cattle entering a CFO carry Campylobacter jejuni, including subtypes resistant to AMAs. The large numbers of cattle arriving from diverse locations at the CFOs and intermingling within the CFOs over time, coupled with the high-density housing of animals, the high rates of transmission of C. jejuni subtypes among animals, and the extensive use of AMAs merge to create an ideal situation where the proliferation of diverse antimicrobial-resistant C. jejuni subtypes is facilitated. Considering that Southern Alberta reports high rates of campylobacteriosis in the human population and that many of these clinical cases are due to C. jejuni subtypes associated with cattle, it is likely that the characteristics of beef cattle CFOs favor the propagation of clinically relevant C. jejuni subtypes, including those resistant to medically important AMAs, which constitute a risk to human health.
Keywords: Campylobacter jejuni; antimicrobial resistance; beef cattle; comparative genomic fingerprinting; population structure; transmission.
© Crown copyright 2018.
Figures


References
-
- Government of Alberta. 2001. Census of agriculture for Alberta, Government of Alberta, Alberta, Canada.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

