DeSUMOylation of MKK7 kinase by the SUMO2/3 protease SENP3 potentiates lipopolysaccharide-induced inflammatory signaling in macrophages
- PMID: 29352108
- PMCID: PMC5857993
- DOI: 10.1074/jbc.M117.816769
DeSUMOylation of MKK7 kinase by the SUMO2/3 protease SENP3 potentiates lipopolysaccharide-induced inflammatory signaling in macrophages
Abstract
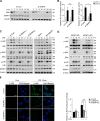
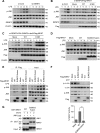
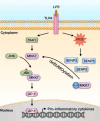
Protein SUMOylation has been reported to play a role in innate immune response, but the enzymes, substrates, and consequences of the specific inflammatory signaling events are largely unknown. Reactive oxygen species (ROS) are abundantly produced during macrophage activation and required for Toll-like receptor 4 (TLR4)-mediated inflammatory signaling. Previously, we demonstrated that SENP3 is a redox-sensitive SUMO2/3 protease. To explore any links between reversible SUMOylation and ROS-related inflammatory signaling in macrophage activation, we generated mice with Senp3 conditional knock-out in myeloid cells. In bacterial lipopolysaccharide (LPS)-induced in vitro and in vivo inflammation models, we found that SENP3 deficiency markedly compromises the activation of TLR4 inflammatory signaling and the production of proinflammatory cytokines in macrophages exposed to LPS. Moreover, Senp3 conditional knock-out mice were significantly less susceptible to septic shock. Of note, SENP3 deficiency was associated with impairment in JNK phosphorylation. We found that MKK7, which selectively phosphorylates JNK, is a SENP3 substrate and that SENP3-mediated deSUMOylation of MKK7 may favor its binding to JNK. Importantly, ROS-dependent SENP3 accumulation and MKK7 deSUMOylation rapidly occurred after LPS stimulation. In conclusion, our findings indicate that SENP3 potentiates LPS-induced TLR4 signaling via deSUMOylation of MKK7 leading to enhancement in JNK phosphorylation and the downstream events. Therefore this work provides novel mechanistic insights into redox regulation of innate immune responses.
Keywords: ROS; SENP3; c-Jun N-terminal kinase (JNK); inflammation; innate immunity; macrophage; sumoylation.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Myeloid SENP3 deficiency protects mice from diet and age-induced obesity via regulation of YAP1 SUMOylation.Cell Mol Life Sci. 2023 Dec 9;81(1):4. doi: 10.1007/s00018-023-05050-w. Cell Mol Life Sci. 2023. PMID: 38070059 Free PMC article.
-
SENP3 in monocytes/macrophages up-regulates tissue factor and mediates lipopolysaccharide-induced acute lung injury by enhancing JNK phosphorylation.J Cell Mol Med. 2020 May;24(10):5454-5462. doi: 10.1111/jcmm.15199. Epub 2020 Mar 31. J Cell Mol Med. 2020. PMID: 32237051 Free PMC article.
-
SENP3 facilitates M1 macrophage polarization via the HIF-1α/PKM2 axis in lipopolysaccharide-induced acute lung injury.Innate Immun. 2023 Jan;29(1-2):25-34. doi: 10.1177/17534259231166212. Epub 2023 Apr 5. Innate Immun. 2023. PMID: 37016838 Free PMC article.
-
SENP3: Cancers and diseases.Biochim Biophys Acta Rev Cancer. 2025 Feb;1880(1):189260. doi: 10.1016/j.bbcan.2025.189260. Epub 2025 Jan 5. Biochim Biophys Acta Rev Cancer. 2025. PMID: 39765284 Review.
-
Caspase-11 non-canonical inflammasome: a critical sensor of intracellular lipopolysaccharide in macrophage-mediated inflammatory responses.Immunology. 2017 Oct;152(2):207-217. doi: 10.1111/imm.12787. Epub 2017 Jul 31. Immunology. 2017. PMID: 28695629 Free PMC article. Review.
Cited by
-
Reformulating Pro-Oxidant Microglia in Neurodegeneration.J Clin Med. 2019 Oct 17;8(10):1719. doi: 10.3390/jcm8101719. J Clin Med. 2019. PMID: 31627485 Free PMC article. Review.
-
SUMO-specific Isopeptidases Tuning Cardiac SUMOylation in Health and Disease.Front Mol Biosci. 2021 Nov 19;8:786136. doi: 10.3389/fmolb.2021.786136. eCollection 2021. Front Mol Biosci. 2021. PMID: 34869605 Free PMC article. Review.
-
The effect of ROS-YAP crosstalk on osteoimmune response orchestrating osteogenesis.Cell Cycle. 2023 Jun;22(11):1391-1405. doi: 10.1080/15384101.2023.2211830. Epub 2023 May 9. Cell Cycle. 2023. PMID: 37161399 Free PMC article.
-
SENP3 affects the expression of PYCR1 to promote bladder cancer proliferation and EMT transformation by deSUMOylation of STAT3.Aging (Albany NY). 2022 Oct 11;14(19):8032-8045. doi: 10.18632/aging.204333. Epub 2022 Oct 11. Aging (Albany NY). 2022. PMID: 36227136 Free PMC article.
-
Myeloid SENP3 deficiency protects mice from diet and age-induced obesity via regulation of YAP1 SUMOylation.Cell Mol Life Sci. 2023 Dec 9;81(1):4. doi: 10.1007/s00018-023-05050-w. Cell Mol Life Sci. 2023. PMID: 38070059 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

