Microbial regulation of the L cell transcriptome
- PMID: 29352262
- PMCID: PMC5775357
- DOI: 10.1038/s41598-017-18079-2
Microbial regulation of the L cell transcriptome
Abstract
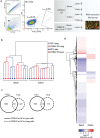
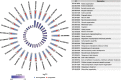
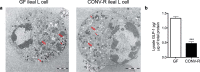
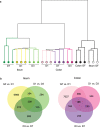
L cells are an important class of enteroendocrine cells secreting hormones such as glucagon like peptide-1 and peptide YY that have several metabolic and physiological effects. The gut is home to trillions of bacteria affecting host physiology, but there has been limited understanding about how the microbiota affects gene expression in L cells. Thus, we rederived the reporter mouse strain, GLU-Venus expressing yellow fluorescent protein under the control of the proglucagon gene, as germ-free (GF). Lpos cells from ileum and colon of GF and conventionally raised (CONV-R) GLU-Venus mice were isolated and subjected to transcriptomic profiling. We observed that the microbiota exerted major effects on ileal L cells. Gene Ontology enrichment analysis revealed that microbiota suppressed biological processes related to vesicle localization and synaptic vesicle cycling in Lpos cells from ileum. This finding was corroborated by electron microscopy of Lpos cells showing reduced numbers of vesicles as well as by demonstrating decreased intracellular GLP-1 content in primary cultures from ileum of CONV-R compared with GF GLU-Venus mice. By analysing Lpos cells following colonization of GF mice we observed that the greatest transcriptional regulation was evident within 1 day of colonization. Thus, the microbiota has a rapid and pronounced effect on the L cell transcriptome, predominantly in the ileum.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

