Functional Characteristics of the Flying Squirrel's Cecal Microbiota under a Leaf-Based Diet, Based on Multiple Meta-Omic Profiling
- PMID: 29354108
- PMCID: PMC5758534
- DOI: 10.3389/fmicb.2017.02622
Functional Characteristics of the Flying Squirrel's Cecal Microbiota under a Leaf-Based Diet, Based on Multiple Meta-Omic Profiling
Abstract
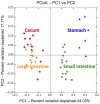
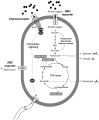
Mammalian herbivores rely on microbial activities in an expanded gut chamber to convert plant biomass into absorbable nutrients. Distinct from ruminants, small herbivores typically have a simple stomach but an enlarged cecum to harbor symbiotic microbes; however, knowledge of this specialized gut structure and characteristics of its microbial contents is limited. Here, we used leaf-eating flying squirrels as a model to explore functional characteristics of the cecal microbiota adapted to a high-fiber, toxin-rich diet. Specifically, environmental conditions across gut regions were evaluated by measuring mass, pH, feed particle size, and metabolomes. Then, parallel metagenomes and metatranscriptomes were used to detect microbial functions corresponding to the cecal environment. Based on metabolomic profiles, >600 phytochemical compounds were detected, although many were present only in the foregut and probably degraded or transformed by gut microbes in the hindgut. Based on metagenomic (DNA) and metatranscriptomic (RNA) profiles, taxonomic compositions of the cecal microbiota were dominated by bacteria of the Firmicutes taxa; they contained major gene functions related to degradation and fermentation of leaf-derived compounds. Based on functional compositions, genes related to multidrug exporters were rich in microbial genomes, whereas genes involved in nutrient importers were rich in microbial transcriptomes. In addition, genes encoding chemotaxis-associated components and glycoside hydrolases specific for plant beta-glycosidic linkages were abundant in both DNA and RNA. This exploratory study provides findings which may help to form molecular-based hypotheses regarding functional contributions of symbiotic gut microbiota in small herbivores with folivorous dietary habits.
Keywords: animal-microbe interaction; ecological adaptation; gut microbiota; metabolomics; metagenomics; metatranscriptomics.
Figures







References
-
- Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. - PubMed
-
- Brulc J. M., Antonopoulos D. A., Miller M. E. B., Wilson M. K., Yannarell A. C., Dinsdale E. A., et al. . (2009). Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 1948–1953. 10.1073/pnas.0806191105 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

