On the Origin of the Non-brittle Rachis Trait of Domesticated Einkorn Wheat
- PMID: 29354137
- PMCID: PMC5758593
- DOI: 10.3389/fpls.2017.02031
On the Origin of the Non-brittle Rachis Trait of Domesticated Einkorn Wheat
Abstract
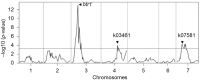
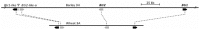
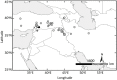
Einkorn and emmer wheat together with barley were among the first cereals domesticated by humans more than 10,000 years ago, long before durum or bread wheat originated. Domesticated einkorn wheat differs from its wild progenitor in basic morphological characters such as the grain dispersal system. This study identified the Non-brittle rachis 1 (btr1) and Non-brittle rachis 2 (btr2) in einkorn as homologous to barley. Re-sequencing of the Btr1 and Btr2 in a collection of 53 lines showed that a single non-synonymous amino acid substitution (alanine to threonine) at position 119 at btr1, is responsible for the non-brittle rachis trait in domesticated einkorn. Tracing this haplotype variation back to wild einkorn samples provides further evidence that the einkorn progenitor came from the Northern Levant. We show that the geographical origin of domesticated haplotype coincides with the non-brittle domesticated barley haplotypes, which suggest the non-brittle rachis phenotypes of einkorn and barley were fixed in same geographic area in today's South-east Turkey.
Keywords: agricultural origins; domestication; einkorn; non-brittle rachis; wheat.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

