Mapping population-based structural connectomes
- PMID: 29355769
- PMCID: PMC5910206
- DOI: 10.1016/j.neuroimage.2017.12.064
Mapping population-based structural connectomes
Abstract
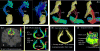
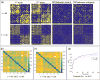
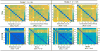
Advances in understanding the structural connectomes of human brain require improved approaches for the construction, comparison and integration of high-dimensional whole-brain tractography data from a large number of individuals. This article develops a population-based structural connectome (PSC) mapping framework to address these challenges. PSC simultaneously characterizes a large number of white matter bundles within and across different subjects by registering different subjects' brains based on coarse cortical parcellations, compressing the bundles of each connection, and extracting novel connection weights. A robust tractography algorithm and streamline post-processing techniques, including dilation of gray matter regions, streamline cutting, and outlier streamline removal are applied to improve the robustness of the extracted structural connectomes. The developed PSC framework can be used to reproducibly extract binary networks, weighted networks and streamline-based brain connectomes. We apply the PSC to Human Connectome Project data to illustrate its application in characterizing normal variations and heritability of structural connectomes in healthy subjects.
Keywords: Brain connectome; Diffusion MRI imaging; Functional principal component analysis; Human connectome project; Population-based structural connectome; Streamline variation decomposition.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



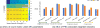



References
-
- Alexander AL, Lee JE, Lazar M, Boudos R, DuBray MB, Oakes TR, Miller JN, Lu J, Jeong EK, McMahon WM, et al. Diffusion tensor imaging of the corpus callosum in Autism. Neuroimage. 2007;34:61–73. - PubMed
-
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic Resonance in Medicine. 2000;44:625–632. - PubMed
-
- Bastiani M, Shah NJ, Goebel R, Roebroeck A. Human cortical connectome reconstruction from diffusion weighted MRI: the effect of tractography algorithm. Neuroimage. 2012;62:1732–1749. - PubMed
-
- Buchanan CR, Pernet CR, Gorgolewski KJ, Storkey AJ, Bastin ME. Test–retest reliability of structural brain networks from diffusion MRI. Neuroimage. 2014;86:231–243. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

