Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells
- PMID: 29355843
- PMCID: PMC5785246
- DOI: 10.1172/JCI97555
Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells
Abstract
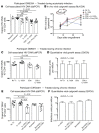
The presence of persistent, latent HIV reservoirs in CD4+ T cells obstructs current efforts to cure infection. The so-called kick-and-kill paradigm proposes to purge these reservoirs by combining latency-reversing agents with immune effectors such as cytotoxic T lymphocytes. Support for this approach is largely based on success in latency models, which do not fully reflect the makeup of latent reservoirs in individuals on long-term antiretroviral therapy (ART). Recent studies have shown that CD8+ T cells have the potential to recognize defective proviruses, which comprise the vast majority of all infected cells, and that the proviral landscape can be shaped over time due to in vivo clonal expansion of infected CD4+ T cells. Here, we have shown that treating CD4+ T cells from ART-treated individuals with combinations of potent latency-reversing agents and autologous CD8+ T cells consistently reduced cell-associated HIV DNA, but failed to deplete replication-competent virus. These CD8+ T cells recognized and potently eliminated CD4+ T cells that were newly infected with autologous reservoir virus, ruling out a role for both immune escape and CD8+ T cell dysfunction. Thus, our results suggest that cells harboring replication-competent HIV possess an inherent resistance to CD8+ T cells that may need to be addressed to cure infection.
Keywords: AIDS/HIV; Drug therapy; Immunology; T cells.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

