Interactome INSIDER: a structural interactome browser for genomic studies
- PMID: 29355848
- PMCID: PMC6026581
- DOI: 10.1038/nmeth.4540
Interactome INSIDER: a structural interactome browser for genomic studies
Abstract
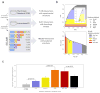
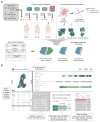
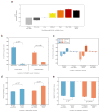
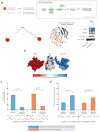
We present Interactome INSIDER, a tool to link genomic variant information with structural protein-protein interactomes. Underlying this tool is the application of machine learning to predict protein interaction interfaces for 185,957 protein interactions with previously unresolved interfaces in human and seven model organisms, including the entire experimentally determined human binary interactome. Predicted interfaces exhibit functional properties similar to those of known interfaces, including enrichment for disease mutations and recurrent cancer mutations. Through 2,164 de novo mutagenesis experiments, we show that mutations of predicted and known interface residues disrupt interactions at a similar rate and much more frequently than mutations outside of predicted interfaces. To spur functional genomic studies, Interactome INSIDER (http://interactomeinsider.yulab.org) enables users to identify whether variants or disease mutations are enriched in known and predicted interaction interfaces at various resolutions. Users may explore known population variants, disease mutations, and somatic cancer mutations, or they may upload their own set of mutations for this purpose.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

