MutScan: fast detection and visualization of target mutations by scanning FASTQ data
- PMID: 29357822
- PMCID: PMC5778627
- DOI: 10.1186/s12859-018-2024-6
MutScan: fast detection and visualization of target mutations by scanning FASTQ data
Abstract
Background: Some types of clinical genetic tests, such as cancer testing using circulating tumor DNA (ctDNA), require sensitive detection of known target mutations. However, conventional next-generation sequencing (NGS) data analysis pipelines typically involve different steps of filtering, which may cause miss-detection of key mutations with low frequencies. Variant validation is also indicated for key mutations detected by bioinformatics pipelines. Typically, this process can be executed using alignment visualization tools such as IGV or GenomeBrowse. However, these tools are too heavy and therefore unsuitable for validating mutations in ultra-deep sequencing data.
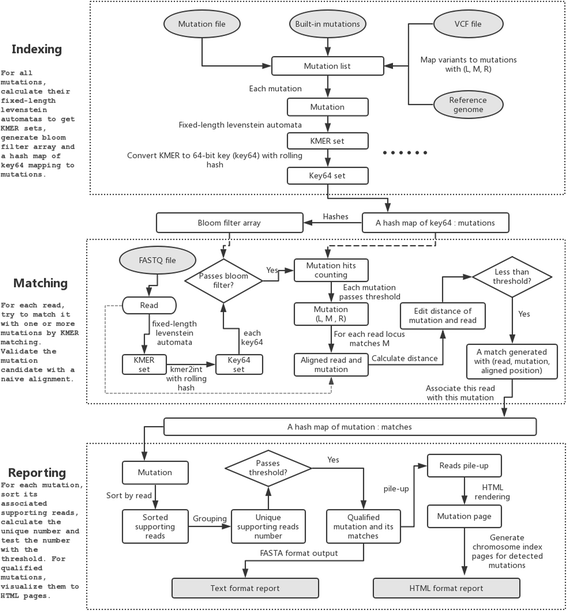
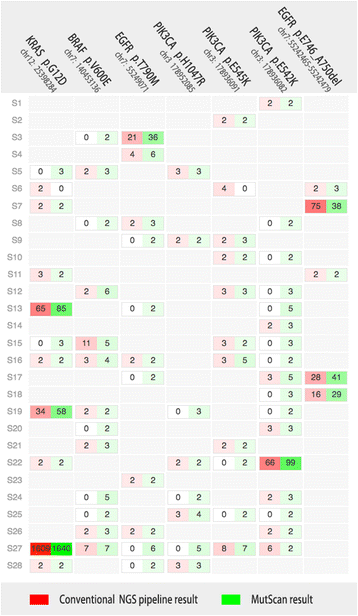
Result: We developed MutScan to address problems of sensitive detection and efficient validation for target mutations. MutScan involves highly optimized string-searching algorithms, which can scan input FASTQ files to grab all reads that support target mutations. The collected supporting reads for each target mutation will be piled up and visualized using web technologies such as HTML and JavaScript. Algorithms such as rolling hash and bloom filter are applied to accelerate scanning and make MutScan applicable to detect or visualize target mutations in a very fast way.
Conclusion: MutScan is a tool for the detection and visualization of target mutations by only scanning FASTQ raw data directly. Compared to conventional pipelines, this offers a very high performance, executing about 20 times faster, and offering maximal sensitivity since it can grab mutations with even one single supporting read. MutScan visualizes detected mutations by generating interactive pile-ups using web technologies. These can serve to validate target mutations, thus avoiding false positives. Furthermore, MutScan can visualize all mutation records in a VCF file to HTML pages for cloud-friendly VCF validation. MutScan is an open source tool available at GitHub: https://github.com/OpenGene/MutScan.
Keywords: Fast detection; MutScan; Mutation scan; Variant visualization.
Conflict of interest statement
Ethics approval and consent to participate
N/A
Consent for publication
N/A
Competing interests
The authors have the following interests: Shifu Chen, Tanxiao Huang, Hong Li and Mingyan Xu are employed by HaploX BioTechnology. There are no patents, products in development or marketed products to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- 61472411/National Natural Science Foundation of China/International
- 2015AA043203/The national 863 Program of China/International
- KC2015JSJS0028A/Technology Development and Creative Design Program of Nanshan Shenzhen/International
- JSGG20160229123927512/Special Funds for Future Industries of Shenzhen/International
- CYZZ20150527145115656/SZSTI Entrepreneurship Funds of Shenzhen/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

