Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep
- PMID: 29357834
- PMCID: PMC5778797
- DOI: 10.1186/s12864-018-4447-x
Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep
Abstract
Background: One of the approaches to detect genetics variants affecting fitness traits is to identify their surrounding genomic signatures of past selection. With established methods for detecting selection signatures and the current and future availability of large datasets, such studies should have the power to not only detect these signatures but also to infer their selective histories. Domesticated animals offer a powerful model for these approaches as they adapted rapidly to environmental and human-mediated constraints in a relatively short time. We investigated this question by studying a large dataset of 542 individuals from 27 domestic sheep populations raised in France, genotyped for more than 500,000 SNPs.
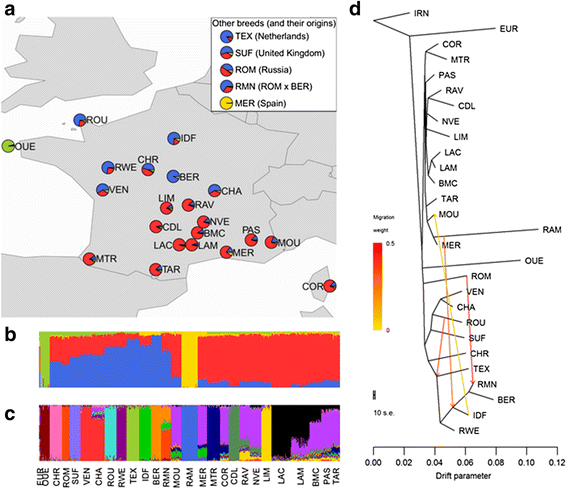
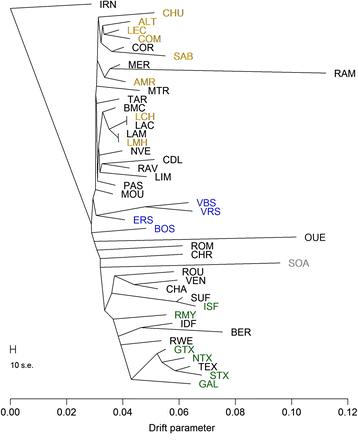
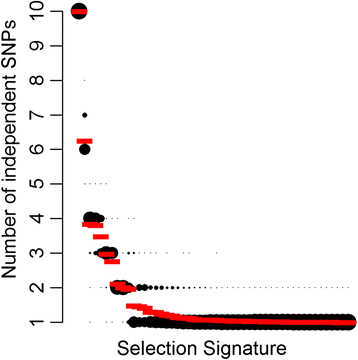
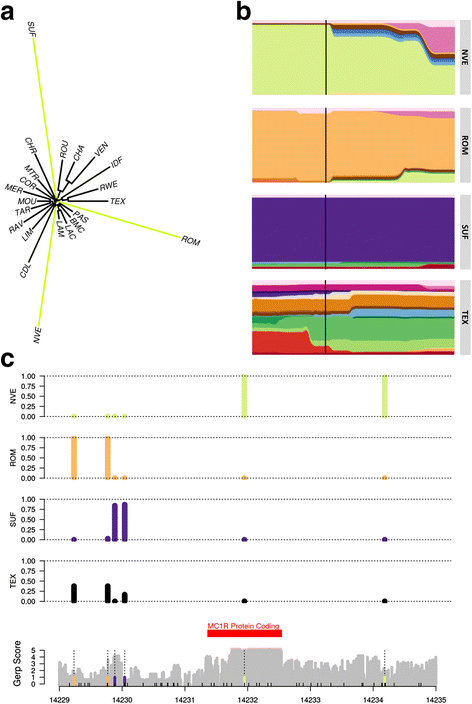
Results: Population structure analysis revealed that this set of populations harbour a large part of European sheep diversity in a small geographical area, offering a powerful model for the study of adaptation. Identification of extreme SNP and haplotype frequency differences between populations listed 126 genomic regions likely affected by selection. These signatures revealed selection at loci commonly identified as selection targets in many species ("selection hotspots") including ABCG2, LCORL/NCAPG, MSTN, and coat colour genes such as ASIP, MC1R, MITF, and TYRP1. For one of these regions (ABCG2, LCORL/NCAPG), we could propose a historical scenario leading to the introgression of an adaptive allele into a new genetic background. Among selection signatures, we found clear evidence for parallel selection events in different genetic backgrounds, most likely for different mutations. We confirmed this allelic heterogeneity in one case by resequencing the MC1R gene in three black-faced breeds.
Conclusions: Our study illustrates how dense genetic data in multiple populations allows the deciphering of evolutionary history of populations and of their adaptive mutations.
Keywords: Adaptation; Allelic heterogeneity; Introgression; Selective sweeps.
Conflict of interest statement
Ethics approval
Not applicable: animals did not belong to any experimental design but were sampled by veterinarians and/or under veterinarian supervision for routine veterinary care.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
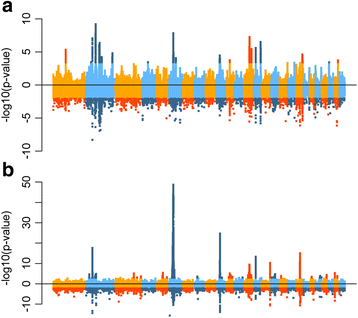
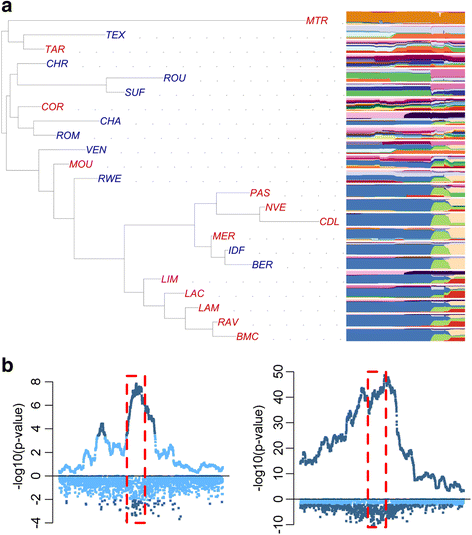
References
-
- Dobney K, Larson G. Genetics and animal domestication: new windows on an elusive process. J Zool. 2006;269:261–271.
-
- Andersson L. How selective sweeps in domestic animals provide new insight into biological mechanisms. J Intern Med. 2012;271:1–14. - PubMed
-
- Kim E-S, Elbeltagy AR, Aboul-Naga AM, Rischkowsky B, Sayre B, Mwacharo JM, et al. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity (Edinb). Nature Publishing Group; 2016;116:1–10. Available from: http://www.nature.com/doifinder/10.1038/hdy.2015.94. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

