Isogenic FUS-eGFP iPSC Reporter Lines Enable Quantification of FUS Stress Granule Pathology that Is Rescued by Drugs Inducing Autophagy
- PMID: 29358088
- PMCID: PMC5857889
- DOI: 10.1016/j.stemcr.2017.12.018
Isogenic FUS-eGFP iPSC Reporter Lines Enable Quantification of FUS Stress Granule Pathology that Is Rescued by Drugs Inducing Autophagy
Abstract
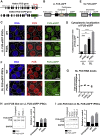
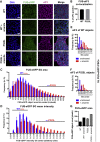
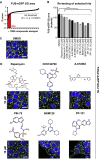
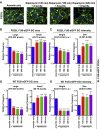
Perturbations in stress granule (SG) dynamics may be at the core of amyotrophic lateral sclerosis (ALS). Since SGs are membraneless compartments, modeling their dynamics in human motor neurons has been challenging, thus hindering the identification of effective therapeutics. Here, we report the generation of isogenic induced pluripotent stem cells carrying wild-type and P525L FUS-eGFP. We demonstrate that FUS-eGFP is recruited into SGs and that P525L profoundly alters their dynamics. With a screening campaign, we demonstrate that PI3K/AKT/mTOR pathway inhibition increases autophagy and ameliorates SG phenotypes linked to P525L FUS by reducing FUS-eGFP recruitment into SGs. Using a Drosophila model of FUS-ALS, we corroborate that induction of autophagy significantly increases survival. Finally, by screening clinically approved drugs for their ability to ameliorate FUS SG phenotypes, we identify a number of brain-penetrant anti-depressants and anti-psychotics that also induce autophagy. These drugs could be repurposed as potential ALS treatments.
Keywords: CRISPR/Cas9n; FUS; amyotrophic lateral sclerosis; autophagy; gene editing; induced pluripotent stem cells; stress granules.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Conte A., Lattante S., Zollino M., Marangi G., Luigetti M., Del Grande A., Servidei S., Trombetta F., Sabatelli M. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul. Disord. 2012;22:73–75. - PubMed
-
- Daigle J.G., Lanson N.A., Jr., Smith R.B., Casci I., Maltare A., Monaghan J., Nichols C.D., Kryndushkin D., Shewmaker F., Pandey U.B. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum. Mol. Genet. 2013;22:1193–1205. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

