Small-Molecule Activators of Protein Phosphatase 2A for the Treatment of Castration-Resistant Prostate Cancer
- PMID: 29358171
- PMCID: PMC5899650
- DOI: 10.1158/0008-5472.CAN-17-0123
Small-Molecule Activators of Protein Phosphatase 2A for the Treatment of Castration-Resistant Prostate Cancer
Abstract
Primary prostate cancer is generally treatable by androgen deprivation therapy, however, later recurrences of castrate-resistant prostate cancer (CRPC) that are more difficult to treat nearly always occur due to aberrant reactivation of the androgen receptor (AR). In this study, we report that CRPC cells are particularly sensitive to the growth-inhibitory effects of reengineered tricyclic sulfonamides, a class of molecules that activate the protein phosphatase PP2A, which inhibits multiple oncogenic signaling pathways. Treatment of CRPC cells with small-molecule activators of PP2A (SMAP) in vitro decreased cellular viability and clonogenicity and induced apoptosis. SMAP treatment also induced an array of significant changes in the phosphoproteome, including most notably dephosphorylation of full-length and truncated isoforms of the AR and downregulation of its regulatory kinases in a dose-dependent and time-dependent manner. In murine xenograft models of human CRPC, the potent compound SMAP-2 exhibited efficacy comparable with enzalutamide in inhibiting tumor formation. Overall, our results provide a preclinical proof of concept for the efficacy of SMAP in AR degradation and CRPC treatment.Significance: A novel class of small-molecule activators of the tumor suppressor PP2A, a serine/threonine phosphatase that inhibits many oncogenic signaling pathways, is shown to deregulate the phosphoproteome and to destabilize the androgen receptor in advanced prostate cancer. Cancer Res; 78(8); 2065-80. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
The Icahn School of Medicine at Mount Sinai on behalf of the authors have filed patents covering composition of matter on the small molecules disclosed herein for the treatment of human cancer and other diseases.
Figures


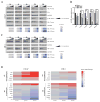

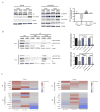


References
-
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. - PubMed
-
- Galsky MD, Small AC, Tsao CK, Oh WK. Clinical development of novel therapeutics for castration-resistant prostate cancer: historic challenges and recent successes. CA Cancer J Clin. 2012;62:299–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

