Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation
- PMID: 29358305
- PMCID: PMC5832932
- DOI: 10.1194/jlr.M081323
Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation
Abstract
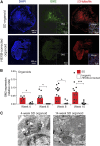
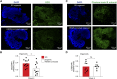
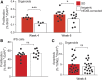
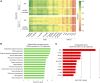
Sandhoff disease, one of the GM2 gangliosidoses, is a lysosomal storage disorder characterized by the absence of β-hexosaminidase A and B activity and the concomitant lysosomal accumulation of its substrate, GM2 ganglioside. It features catastrophic neurodegeneration and death in early childhood. How the lysosomal accumulation of ganglioside might affect the early development of the nervous system is not understood. Recently, cerebral organoids derived from induced pluripotent stem (iPS) cells have illuminated early developmental events altered by disease processes. To develop an early neurodevelopmental model of Sandhoff disease, we first generated iPS cells from the fibroblasts of an infantile Sandhoff disease patient, then corrected one of the mutant HEXB alleles in those iPS cells using CRISPR/Cas9 genome-editing technology, thereby creating isogenic controls. Next, we used the parental Sandhoff disease iPS cells and isogenic HEXB-corrected iPS cell clones to generate cerebral organoids that modeled the first trimester of neurodevelopment. The Sandhoff disease organoids, but not the HEXB-corrected organoids, accumulated GM2 ganglioside and exhibited increased size and cellular proliferation compared with the HEXB-corrected organoids. Whole-transcriptome analysis demonstrated that development was impaired in the Sandhoff disease organoids, suggesting that alterations in neuronal differentiation may occur during early development in the GM2 gangliosidoses.
Keywords: Clustered Regularly Interspaced Short Palindromic Repeats/Cas9; GM2 gangliosidosis; Tay-Sachs disease; brain development; brain lipids; gangliosides; patient-derived induced pluripotent stem cells; sphingolipids; storage diseases.
Figures





References
-
- Sango K., Yamanaka S., Hoffmann A., Okuda Y., Grinberg A., Westphal H., McDonald M. P., Crawley J. N., Sandhoff K., Suzuki K., et al. . 1995. Mouse models of Tay-Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat. Genet. 11: 170–176. - PubMed
-
- Huang J. Q., Trasler J. M., Igdoura S., Michaud J., Hanal N., and Gravel R. A.. 1997. Apoptotic cell death in mouse models of GM2 gangliosidosis and observations on human Tay-Sachs and Sandhoff diseases. Hum. Mol. Genet. 6: 1879–1885. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

