Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs
- PMID: 29358381
- PMCID: PMC5819447
- DOI: 10.1073/pnas.1718910115
Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs
Abstract
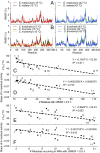
Orthologous proteins of species adapted to different temperatures exhibit differences in stability and function that are interpreted to reflect adaptive variation in structural "flexibility." However, quantifying flexibility and comparing flexibility across proteins has remained a challenge. To address this issue, we examined temperature effects on cytosolic malate dehydrogenase (cMDH) orthologs from differently thermally adapted congeners of five genera of marine molluscs whose field body temperatures span a range of ∼60 °C. We describe consistent patterns of convergent evolution in adaptation of function [temperature effects on KM of cofactor (NADH)] and structural stability (rate of heat denaturation of activity). To determine how these differences depend on flexibilities of overall structure and of regions known to be important in binding and catalysis, we performed molecular dynamics simulation (MDS) analyses. MDS analyses revealed a significant negative correlation between adaptation temperature and heat-induced increase of backbone atom movements [root mean square deviation (rmsd) of main-chain atoms]. Root mean square fluctuations (RMSFs) of movement by individual amino acid residues varied across the sequence in a qualitatively similar pattern among orthologs. Regions of sequence involved in ligand binding and catalysis-termed mobile regions 1 and 2 (MR1 and MR2), respectively-showed the largest values for RMSF. Heat-induced changes in RMSF values across the sequence and, importantly, in MR1 and MR2 were greatest in cold-adapted species. MDS methods are shown to provide powerful tools for examining adaptation of enzymes by providing a quantitative index of protein flexibility and identifying sequence regions where adaptive change in flexibility occurs.
Keywords: adaptation; amino acid sequence; evolution; malate dehydrogenase; molecular dynamics simulations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Somero GN, Lockwood BL, Tomanek L. Biochemical Adaptation: Response to Environmental Challenges from Life’s Origins to the Anthropocene. Sinauer Associates; Sunderland, MA: 2017.
-
- Fields PA, Dong Y, Meng X, Somero GN. Adaptations of protein structure and function to temperature: There is more than one way to ‘skin a cat’. J Exp Biol. 2015;218:1801–1811. - PubMed
-
- Feller G. Enzyme function at low temperatures in psychrophiles. In: Siddiqui KS, Thomas T, editors. Protein Adaptation in Extremophiles. Nova Science; Hauppauge, NY: 2008. pp. 35–69.
-
- Feller G, Gerday C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat Rev Microbiol. 2003;1:200–208. - PubMed
-
- Dalhus B, et al. Structural basis for thermophilic protein stability: Structures of thermophilic and mesophilic malate dehydrogenases. J Mol Biol. 2002;318:707–721. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

