Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes
- PMID: 29358391
- PMCID: PMC5819426
- DOI: 10.1073/pnas.1716667115
Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes
Erratum in
-
Correction for Adam et al., Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes.Proc Natl Acad Sci U S A. 2018 Jun 19;115(25):E5836-E5837. doi: 10.1073/pnas.1807540115. Epub 2018 Jun 11. Proc Natl Acad Sci U S A. 2018. PMID: 29891695 Free PMC article. No abstract available.
Abstract
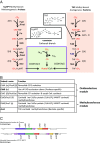
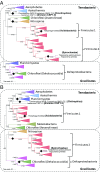
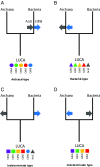
Carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) is a five-subunit enzyme complex responsible for the carbonyl branch of the Wood-Ljungdahl (WL) pathway, considered one of the most ancient metabolisms for anaerobic carbon fixation, but its origin and evolutionary history have been unclear. While traditionally associated with methanogens and acetogens, the presence of CODH/ACS homologs has been reported in a large number of uncultured anaerobic lineages. Here, we have carried out an exhaustive phylogenomic study of CODH/ACS in over 6,400 archaeal and bacterial genomes. The identification of complete and likely functional CODH/ACS complexes in these genomes significantly expands its distribution in microbial lineages. The CODH/ACS complex displays astounding conservation and vertical inheritance over geological times. Rare intradomain and interdomain transfer events might tie into important functional transitions, including the acquisition of CODH/ACS in some archaeal methanogens not known to fix carbon, the tinkering of the complex in a clade of model bacterial acetogens, or emergence of archaeal-bacterial hybrid complexes. Once these transfers were clearly identified, our results allowed us to infer the presence of a CODH/ACS complex with at least four subunits in the last universal common ancestor (LUCA). Different scenarios on the possible role of ancestral CODH/ACS are discussed. Despite common assumptions, all are equally compatible with an autotrophic, mixotrophic, or heterotrophic LUCA. Functional characterization of CODH/ACS from a larger spectrum of bacterial and archaeal lineages and detailed evolutionary analysis of the WL methyl branch will help resolve this issue.
Keywords: LUCA; Wood–Ljungdahl pathway; acetogens; evolution; methanogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Fuchs G. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–658. - PubMed
-
- Peretó J. Out of fuzzy chemistry: From prebiotic chemistry to metabolic networks. Chem Soc Rev. 2012;41:5394–5403. - PubMed
-
- Berg IA, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. - PubMed
-
- Möller-Zinkhan D, Thauer RK. Anaerobic lactate oxidation to 3 CO2 by Archaeoglobus fulgidus via the carbon monoxide dehydrogenase pathway: Demonstration of the acetyl-CoA carbon-carbon cleavage reaction in cell extracts. Arch Microbiol. 1990;153:215–218.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

