eIF2α phosphorylation is pathognomonic for immunogenic cell death
- PMID: 29358668
- PMCID: PMC6113215
- DOI: 10.1038/s41418-017-0044-9
eIF2α phosphorylation is pathognomonic for immunogenic cell death
Abstract
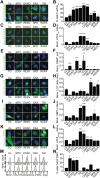
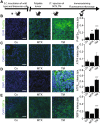
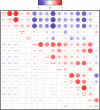
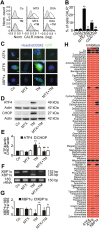
The phosphorylation of eIF2α is essential for the endoplasmic reticulum (ER) stress response, the formation of stress granules, as well as macroautophagy. Several successful anticancer chemotherapeutics have the property to induce immunogenic cell death (ICD), thereby causing anticancer immune responses. ICD is accompanied by the translocation of calreticulin (CALR) from the ER lumen to the plasma membrane, which facilitates the transfer of tumor-associated antigens to dendritic cells. Here we systematically investigated the capacity of anticancer chemotherapeutics to induce signs of ER stress. ICD inducers including anthracyclines and agents that provoke tetraploidization were highly efficient in enhancing the phosphorylation of eIF2α, yet failed to stimulate other signs of ER stress including the transcriptional activation of activating transcription factor 4 (ATF4), the alternative splicing of X-box binding protein 1 (XBP1s) mRNA and the proteolytic cleavage of activating transcription factor 6 (ATF6) both in vitro and in cancers established in mice. Systematic analyses of clinically used anticancer chemotherapeutics revealed that only eIF2α phosphorylation, but none of the other signs of ER stress, correlated with CALR exposure. eIF2α phosphorylation induced by mitoxantrone, a prototype ICD-inducing anthracyline, was mediated by eIF2α kinase-3 (EIF2AK3). Machine-learning approaches were used to determine the physicochemical properties of drugs that induce ICD, revealing that the sole ER stress response relevant to the algorithm is eIF2α phosphorylation with its downstream consequences CALR exposure, stress granule formation and autophagy induction. Importantly, this approach could reduce the complexity of compound libraries to identify ICD inducers based on their physicochemical and structural characteristics. In summary, it appears that eIF2α phosphorylation constitutes a pathognomonic characteristic of ICD.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

