Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans
- PMID: 29358710
- PMCID: PMC5778133
- DOI: 10.1038/s41467-017-02235-3
Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans
Abstract
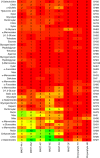
Single-celled eukaryotes (protists) are critical players in global biogeochemical cycling of nutrients and energy in the oceans. While their roles as primary producers and grazers are well appreciated, other aspects of their life histories remain obscure due to challenges in culturing and sequencing their natural diversity. Here, we exploit single-cell genomics and metagenomics data from the circumglobal Tara Oceans expedition to analyze the genome content and apparent oceanic distribution of seven prevalent lineages of uncultured heterotrophic stramenopiles. Based on the available data, each sequenced genome or genotype appears to have a specific oceanic distribution, principally correlated with water temperature and depth. The genome content provides hypotheses for specialization in terms of cell motility, food spectra, and trophic stages, including the potential impact on their lifestyles of horizontal gene transfer from prokaryotes. Our results support the idea that prominent heterotrophic marine protists perform diverse functions in ocean ecology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

