An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes
- PMID: 29359744
- PMCID: PMC5784781
- DOI: 10.1038/nchem.2908
An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes
Abstract
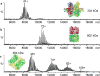
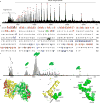
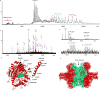
Mass spectrometry (MS) has become a crucial technique for the analysis of protein complexes. Native MS has traditionally examined protein subunit arrangements, while proteomics MS has focused on sequence identification. These two techniques are usually performed separately without taking advantage of the synergies between them. Here we describe the development of an integrated native MS and top-down proteomics method using Fourier-transform ion cyclotron resonance (FTICR) to analyse macromolecular protein complexes in a single experiment. We address previous concerns of employing FTICR MS to measure large macromolecular complexes by demonstrating the detection of complexes up to 1.8 MDa, and we demonstrate the efficacy of this technique for direct acquirement of sequence to higher-order structural information with several large complexes. We then summarize the unique functionalities of different activation/dissociation techniques. The platform expands the ability of MS to integrate proteomics and structural biology to provide insights into protein structure, function and regulation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
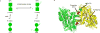



References
-
- Sharon M. How Far Can We Go with Structural Mass Spectrometry of Protein Complexes? J Am Soc Mass Spectrom. 2010;21:487–500. - PubMed
-
- Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Meth. 2008;5:927–933. - PubMed
-
- Lorenzen K, Duijn EV. Current Protocols in Protein Science. John Wiley & Sons, Inc; 2001.
-
- van Duijn E. Current Limitations in Native Mass Spectrometry Based Structural Biology. J Am Soc Mass Spectrom. 2010;21:971–978. - PubMed
-
- Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein Complexes in the Gas Phase: Technology for Structural Genomics and Proteomics. Chem Rev. 2007;107:3544–3567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

