Responders and non-responders to influenza vaccination: A DNA methylation approach on blood cells
- PMID: 29360511
- PMCID: PMC5989724
- DOI: 10.1016/j.exger.2018.01.019
Responders and non-responders to influenza vaccination: A DNA methylation approach on blood cells
Abstract
Several evidences indicate that aging negatively affects the effectiveness of influenza vaccination. Although it is well established that immunosenescence has an important role in vaccination response, the molecular pathways underlying this process are largely unknown. Given the importance of epigenetic remodeling in aging, here we analyzed the relationship between responsiveness to influenza vaccination and DNA methylation profiles in healthy subjects of different ages. Peripheral blood mononuclear cells were collected from 44 subjects (age range: 19-90 years old) immediately before influenza vaccination. Subjects were subsequently classified as responders or non-responders according to hemagglutination inhibition assay 4-6 weeks after the vaccination. Baseline whole genome DNA methylation in peripheral blood mononuclear cells was analyzed using the Illumina® Infinium 450 k microarray. Differential methylation analysis between the two groups (responders and non-responders) was performed through an analysis of variance, correcting for age, sex and batch. We identified 83 CpG sites having a nominal p-value <.001 and absolute difference in DNA methylation of at least 0.05 between the two groups. For some CpG sites, we observed age-dependent decrease or increase in methylation, which in some cases was specific for the responders and non-responders groups. Finally, we divided the cohort in two subgroups including younger (age < 50) and older (age ≥ 50) subjects and compared DNA methylation between responders and non-responders, correcting for sex and batch in each subgroup. We identified 142 differentially methylated CpG sites in the young subgroup and 305 in the old subgroup, suggesting a larger epigenetic remodeling at older ages. Interestingly, some of the differentially methylated probes mapped in genes involved in immunosenescence (CD40) and in innate immunity responses (CXCL16, ULK1, BCL11B, BTC). In conclusion, the analysis of epigenetic landscape can shed light on the biological basis of vaccine responsiveness during aging, possibly providing new appropriate biomarkers of this process.
Keywords: Aging; DNA methylation; Immunosenescence; Influenza vaccination.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

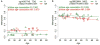
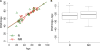
References
-
- Al-Yahya S, Mahmoud L, Al-Zoghaibi F, Al-Tuhami A, Amer H, Almajhdi FN, Polyak SJ, Khabar KSA. Human Cytokinome Analysis for Interferon Response. J. Virol. 2015;89:7108–7119. https://doi.org/10.1128/JVI.03729-14. - DOI - PMC - PubMed
-
- Arts RJW, Joosten LAB, Netea MG. Immunometabolic circuits in trained immunity. Semin. Immunol. 2016;28:425–430. https://doi.org/10.1016/j.smim.2016.09.002. - DOI - PubMed
-
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinforma. Oxf. Engl. 2014;30:1363–1369. https://doi.org/10.1093/bioinformatics/btu049. - DOI - PMC - PubMed
-
- Bacalini MG, Boattini A, Gentilini D, Giampieri E, Pirazzini C, Giuliani C, Fontanesi E, Remondini D, Capri M, Del Rio A, Luiselli D, Vitale G, Mari D, Castellani G, Di Blasio AM, Salvioli S, Franceschi C, Garagnani P. A meta-analysis on age-associated changes in blood DNA methylation: results from an original analysis pipeline for Infinium 450k data. Aging. 2015;7:97–109. https://doi.org/10.18632/aging.100718. - DOI - PMC - PubMed
-
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan J-B, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. https://doi.org/10.1016/j.ygeno.2011.07.007. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

