Identification and analysis of OsttaDSP, a phosphoglucan phosphatase from Ostreococcus tauri
- PMID: 29360855
- PMCID: PMC5779698
- DOI: 10.1371/journal.pone.0191621
Identification and analysis of OsttaDSP, a phosphoglucan phosphatase from Ostreococcus tauri
Abstract
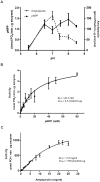
Ostreococcus tauri, the smallest free-living (non-symbiotic) eukaryote yet described, is a unicellular green alga of the Prasinophyceae family. It has a very simple cellular organization and presents a unique starch granule and chloroplast. However, its starch metabolism exhibits a complexity comparable to higher plants, with multiple enzyme forms for each metabolic reaction. Glucan phosphatases, a family of enzymes functionally conserved in animals and plants, are essential for normal starch or glycogen degradation in plants and mammals, respectively. Despite the importance of O. tauri microalgae in evolution, there is no information available concerning the enzymes involved in reversible phosphorylation of starch. Here, we report the molecular cloning and heterologous expression of the gene coding for a dual specific phosphatase from O. tauri (OsttaDSP), homologous to Arabidopsis thaliana LSF2. The recombinant enzyme was purified to electrophoretic homogeneity to characterize its oligomeric and kinetic properties accurately. OsttaDSP is a homodimer of 54.5 kDa that binds and dephosphorylates amylopectin. Also, we also determined that residue C162 is involved in catalysis and possibly also in structural stability of the enzyme. Our results could contribute to better understand the role of glucan phosphatases in the metabolism of starch in green algae.
Conflict of interest statement
Figures




References
-
- Tester R, Karkalas J, Qi X. Starch structure and digestibility enzyme-substrate relationship. World’s Poultry Science Journal. 2004;60(02):186–95.
-
- Blennow A, Bay-Smidt AM, Olsen CE, Møller BL. The distribution of covalently bound phosphate in the starch granule in relation to starch crystallinity. International Journal of Biological Macromolecules. 2000;27(3):211–8. - PubMed
-
- Carciofi M, Shaik SS, Jensen SL, Blennow A, Svensson JT, Vincze É, et al. Hyperphosphorylation of cereal starch. Journal of Cereal Science. 2011;54(3):339–46.
-
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an α-glucan, water dikinase. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):7166–71. doi: 10.1073/pnas.062053099 - DOI - PMC - PubMed
-
- Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS letters. 2006;580(20):4872–6. Epub 2006/08/18. doi: 10.1016/j.febslet.2006.07.085 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

