The human cytomegalovirus terminase complex as an antiviral target: a close-up view
- PMID: 29361041
- PMCID: PMC5972660
- DOI: 10.1093/femsre/fuy004
The human cytomegalovirus terminase complex as an antiviral target: a close-up view
Abstract
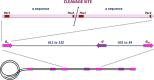
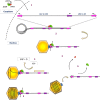
Human cytomegalovirus (HCMV) is responsible for life-threatening infections in immunocompromised individuals and can cause serious congenital malformations. Available antivirals target the viral polymerase but are subject to cross-resistance and toxicity. New antivirals targeting other replication steps and inducing fewer adverse effects are therefore needed. During HCMV replication, DNA maturation and packaging are performed by the terminase complex, which cleaves DNA to package the genome into the capsid. Identified in herpesviruses and bacteriophages, and with no counterpart in mammalian cells, these terminase proteins are ideal targets for highly specific antivirals. A new terminase inhibitor, letermovir, recently proved effective against HCMV in phase III clinical trials, but the mechanism of action is unclear. Letermovir has no significant activity against other herpesvirus or non-human CMV. This review focuses on the highly conserved mechanism of HCMV DNA-packaging and the potential of the terminase complex to serve as an antiviral target. We describe the intrinsic mechanism of DNA-packaging, highlighting the structure-function relationship of HCMV terminase complex components.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

