SemEHR: A general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research
- PMID: 29361077
- PMCID: PMC6019046
- DOI: 10.1093/jamia/ocx160
SemEHR: A general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research
Abstract
Objective: Unlocking the data contained within both structured and unstructured components of electronic health records (EHRs) has the potential to provide a step change in data available for secondary research use, generation of actionable medical insights, hospital management, and trial recruitment. To achieve this, we implemented SemEHR, an open source semantic search and analytics tool for EHRs.
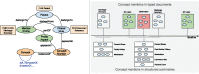
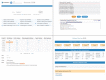
Methods: SemEHR implements a generic information extraction (IE) and retrieval infrastructure by identifying contextualized mentions of a wide range of biomedical concepts within EHRs. Natural language processing annotations are further assembled at the patient level and extended with EHR-specific knowledge to generate a timeline for each patient. The semantic data are serviced via ontology-based search and analytics interfaces.
Results: SemEHR has been deployed at a number of UK hospitals, including the Clinical Record Interactive Search, an anonymized replica of the EHR of the UK South London and Maudsley National Health Service Foundation Trust, one of Europe's largest providers of mental health services. In 2 Clinical Record Interactive Search-based studies, SemEHR achieved 93% (hepatitis C) and 99% (HIV) F-measure results in identifying true positive patients. At King's College Hospital in London, as part of the CogStack program (github.com/cogstack), SemEHR is being used to recruit patients into the UK Department of Health 100 000 Genomes Project (genomicsengland.co.uk). The validation study suggests that the tool can validate previously recruited cases and is very fast at searching phenotypes; time for recruitment criteria checking was reduced from days to minutes. Validated on open intensive care EHR data, Medical Information Mart for Intensive Care III, the vital signs extracted by SemEHR can achieve around 97% accuracy.
Conclusion: Results from the multiple case studies demonstrate SemEHR's efficiency: weeks or months of work can be done within hours or minutes in some cases. SemEHR provides a more comprehensive view of patients, bringing in more and unexpected insight compared to study-oriented bespoke IE systems. SemEHR is open source, available at https://github.com/CogStack/SemEHR.
Figures

References
Publication types
MeSH terms
Grants and funding
- BHF_/British Heart Foundation/United Kingdom
- MR/S004149/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_14089/MRC_/Medical Research Council/United Kingdom
- MR/K006584/1/WT_/Wellcome Trust/United Kingdom
- ARC_/Arthritis Research UK/United Kingdom
- MR/L014815/1/MRC_/Medical Research Council/United Kingdom
- CSO_/Chief Scientist Office/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- MC_PC_17214/MRC_/Medical Research Council/United Kingdom
- 109823/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_14089 /MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

