Drosophila Sidekick is required in developing photoreceptors to enable visual motion detection
- PMID: 29361567
- PMCID: PMC5818003
- DOI: 10.1242/dev.158246
Drosophila Sidekick is required in developing photoreceptors to enable visual motion detection
Abstract
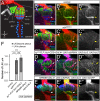
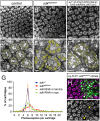
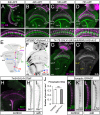
The assembly of functional neuronal circuits requires growth cones to extend in defined directions and recognize the correct synaptic partners. Homophilic adhesion between vertebrate Sidekick proteins promotes synapse formation between retinal neurons involved in visual motion detection. We show here that Drosophila Sidekick accumulates in specific synaptic layers of the developing motion detection circuit and is necessary for normal optomotor behavior. Sidekick is required in photoreceptors, but not in their target lamina neurons, to promote the alignment of lamina neurons into columns and subsequent sorting of photoreceptor axons into synaptic modules based on their precise spatial orientation. Sidekick is also localized to the dendrites of the direction-selective T4 and T5 cells, and is expressed in some of their presynaptic partners. In contrast to its vertebrate homologs, Sidekick is not essential for T4 and T5 to direct their dendrites to the appropriate layers or to receive synaptic contacts. These results illustrate a conserved requirement for Sidekick proteins in establishing visual motion detection circuits that is achieved through distinct cellular mechanisms in Drosophila and vertebrates.
Keywords: Lamina; Optomotor behavior; Photoreceptor; Sidekick; Visual motion detection.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K. L. et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, 731-743. 10.1534/genetics.111.126995 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

