TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency
- PMID: 29361574
- PMCID: PMC5869011
- DOI: 10.1242/dev.161075
TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency
Abstract
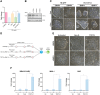
Human embryonic stem cells (hESCs) are exquisitely sensitive to WNT ligands, which rapidly cause differentiation. Therefore, hESC self-renewal requires robust mechanisms to keep the cells in a WNT inactive but responsive state. How they achieve this is largely unknown. We explored the role of transcriptional regulators of WNT signaling, the TCF/LEFs. As in mouse ESCs, TCF7L1 is the predominant family member expressed in hESCs. Genome-wide, it binds a gene cohort involved in primitive streak formation at gastrulation, including NODAL, BMP4 and WNT3 Comparing TCF7L1-bound sites with those bound by the WNT signaling effector β-catenin indicates that TCF7L1 acts largely on the WNT signaling pathway. TCF7L1 overlaps less with the pluripotency regulators OCT4 and NANOG than in mouse ESCs. Gain- and loss-of-function studies indicate that TCF7L1 suppresses gene cohorts expressed in the primitive streak. Interestingly, we find that BMP4, another driver of hESC differentiation, downregulates TCF7L1, providing a mechanism of BMP and WNT pathway intersection. Together, our studies indicate that TCF7L1 plays a major role in maintaining hESC pluripotency, which has implications for human development during gastrulation.
Keywords: BMP4; Gastrulation; Human ES cells; Primitive streak; TCF7L1; WNT signaling.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
Characterization of a chromatin-associated TCF7L1 complex in human embryonic stem cells.Proteomics. 2024 Oct;24(20):e2300641. doi: 10.1002/pmic.202300641. Epub 2024 Apr 17. Proteomics. 2024. PMID: 38629187
-
Tcf7l1 prepares epiblast cells in the gastrulating mouse embryo for lineage specification.Development. 2013 Apr;140(8):1665-75. doi: 10.1242/dev.087387. Epub 2013 Mar 13. Development. 2013. PMID: 23487311 Free PMC article.
-
Tbl1 promotes Wnt-β-catenin signaling-induced degradation of the Tcf7l1 protein in mouse embryonic stem cells.J Cell Sci. 2024 May 1;137(9):jcs261241. doi: 10.1242/jcs.261241. Epub 2024 May 16. J Cell Sci. 2024. PMID: 38639717
-
β-Catenin in pluripotency: adhering to self-renewal or Wnting to differentiate?Int Rev Cell Mol Biol. 2014;312:53-78. doi: 10.1016/B978-0-12-800178-3.00002-6. Int Rev Cell Mol Biol. 2014. PMID: 25262238 Review.
-
Mechanisms of Wnt signaling and control.Wiley Interdiscip Rev Syst Biol Med. 2018 Sep;10(5):e1422. doi: 10.1002/wsbm.1422. Epub 2018 Mar 30. Wiley Interdiscip Rev Syst Biol Med. 2018. PMID: 29600540 Free PMC article. Review.
Cited by
-
TCF3 alternative splicing controlled by hnRNP H/F regulates E-cadherin expression and hESC pluripotency.Genes Dev. 2018 Sep 1;32(17-18):1161-1174. doi: 10.1101/gad.316984.118. Epub 2018 Aug 16. Genes Dev. 2018. PMID: 30115631 Free PMC article.
-
Expression of targets of the RNA-binding protein AUF-1 in human airway epithelium indicates its role in cellular senescence and inflammation.Front Immunol. 2023 Jul 7;14:1192028. doi: 10.3389/fimmu.2023.1192028. eCollection 2023. Front Immunol. 2023. PMID: 37483631 Free PMC article.
-
Host T Cell Dedifferentiation Effects Drive HIV-1 Latency Stability.J Virol. 2022 Mar 9;96(5):e0197421. doi: 10.1128/jvi.01974-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019721 Free PMC article.
-
Aging: A cell source limiting factor in tissue engineering.World J Stem Cells. 2019 Oct 26;11(10):787-802. doi: 10.4252/wjsc.v11.i10.787. World J Stem Cells. 2019. PMID: 31692986 Free PMC article. Review.
-
Role of Mesenchymal Stem Cells in Counteracting Oxidative Stress-Related Neurodegeneration.Int J Mol Sci. 2020 May 7;21(9):3299. doi: 10.3390/ijms21093299. Int J Mol Sci. 2020. PMID: 32392722 Free PMC article. Review.
References
-
- Atlasi Y., Noori R., Gaspar C., Franken P., Sacchetti A., Rafati H., Mahmoudi T., Decraene C., Calin G. A., Merrill B. J. et al. (2013). Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet. 9, e1003424 10.1371/journal.pgen.1003424 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

