Affinity Purification and Comparative Biosensor Analysis of Citrulline-Peptide-Specific Antibodies in Rheumatoid Arthritis
- PMID: 29361749
- PMCID: PMC5796268
- DOI: 10.3390/ijms19010326
Affinity Purification and Comparative Biosensor Analysis of Citrulline-Peptide-Specific Antibodies in Rheumatoid Arthritis
Abstract
Background: In rheumatoid arthritis (RA), anti-citrullinated protein/peptide antibodies (ACPAs) are responsible for disease onset and progression, however, our knowledge is limited on ligand binding affinities of autoantibodies with different citrulline-peptide specificity.
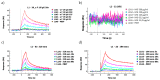
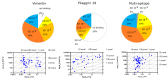
Methods: Citrulline-peptide-specific ACPA IgGs were affinity purified and tested by ELISA. Binding affinities of ACPA IgGs and serum antibodies were compared by surface plasmon resonance (SPR) analysis. Bifunctional nanoparticles harboring a multi-epitope citrulline-peptide and a complement-activating peptide were used to induce selective depletion of ACPA-producing B cells.
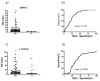
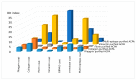
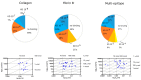
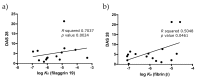
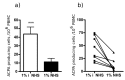
Results: KD values of affinity-purified ACPA IgGs varied between 10-6 and 10-8 M and inversely correlated with disease activity. Based on their cross-reaction with citrulline-peptides, we designed a novel multi-epitope peptide, containing Cit-Gly and Ala-Cit motifs in two-two copies, separated with a short, neutral spacer. This peptide detected antibodies in RA sera with 66% sensitivity and 98% specificity in ELISA and was recognized by 90% of RA sera, while none of the healthy samples in SPR. When coupled to nanoparticles, the multi-epitope peptide specifically targeted and depleted ACPA-producing B cells ex vivo.
Conclusions: The unique multi-epitope peptide designed based on ACPA cross-reactivity might be suitable to develop better diagnostics and novel therapies for RA.
Keywords: affinity; autoantibody; citrulline-peptides; cross-reaction; rheumatoid arthritis; targeting.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

References
-
- Tarcsa E., Marekov L.N., Mei G., Melino G., Lee S.C., Steinert P.M. Protein Unfolding by Peptidylarginine Deiminase. Substrate Specificity and Structural Relationships of the Natural Substrates Trichohyalin and Filaggrin. J. Biol. Chem. 1996;271:30709–30716. doi: 10.1074/jbc.271.48.30709. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

