Down Syndrome Critical Region 1 Gene, Rcan1, Helps Maintain a More Fused Mitochondrial Network
- PMID: 29362227
- PMCID: PMC5924463
- DOI: 10.1161/CIRCRESAHA.117.311522
Down Syndrome Critical Region 1 Gene, Rcan1, Helps Maintain a More Fused Mitochondrial Network
Abstract
Rationale: The regulator of calcineurin 1 (RCAN1) inhibits CN (calcineurin), a Ca2+-activated protein phosphatase important in cardiac remodeling. In humans, RCAN1 is located on chromosome 21 in proximity to the Down syndrome critical region. The hearts and brains of Rcan1 KO mice are more susceptible to damage from ischemia/reperfusion (I/R); however, the underlying cause is not known.
Objective: Mitochondria are key mediators of I/R damage. The goal of these studies was to determine the impact of RCAN1 on mitochondrial dynamics and function.
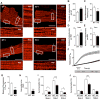
Methods and results: Using both neonatal and isolated adult cardiomyocytes, we show that, when RCAN1 is depleted, the mitochondrial network is more fragmented because of increased CN-dependent activation of the fission protein, DRP1 (dynamin-1-like). Mitochondria in RCAN1-depleted cardiomyocytes have reduced membrane potential, O2 consumption, and generation of reactive oxygen species, as well as a reduced capacity for mitochondrial Ca2+ uptake. RCAN1-depleted cardiomyocytes were more sensitive to I/R; however, pharmacological inhibition of CN, DRP1, or CAPN (calpains; Ca2+-activated proteases) restored protection, suggesting that in the absence of RCAN1, CAPN-mediated damage after I/R is greater because of a decrease in the capacity of mitochondria to buffer cytoplasmic Ca2+. Increasing RCAN1 levels by adenoviral infection was sufficient to enhance fusion and confer protection from I/R. To examine the impact of more modest, and biologically relevant, increases in RCAN1, we compared the mitochondrial network in induced pluripotent stem cells derived from individuals with Down syndrome to that of isogenic, disomic controls. Mitochondria were more fused, and O2 consumption was greater in the trisomic induced pluripotent stem cells; however, coupling efficiency and metabolic flexibility were compromised compared with disomic induced pluripotent stem cells. Depletion of RCAN1 from trisomic induced pluripotent stem cells was sufficient to normalize mitochondrial dynamics and function.
Conclusions: RCAN1 helps maintain a more interconnected mitochondrial network, and maintaining appropriate RCAN1 levels is important to human health and disease.
Keywords: Down syndrome; calcineurin; calpain; ischemia reperfusion injury; mitochondria; mitochondrial dynamics.
© 2018 American Heart Association, Inc.
Figures



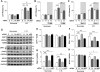



Comment in
-
RCAN1-Calcineurin Axis and the Set-Point for Myocardial Damage During Ischemia-Reperfusion.Circ Res. 2018 Mar 16;122(6):796-798. doi: 10.1161/CIRCRESAHA.118.312787. Circ Res. 2018. PMID: 29700078 Free PMC article. No abstract available.
References
-
- Mozaffarian D, Benjamin EJ, Go AS, et al. Heart Disease and Stroke Statistics—2015 Update. Circulation. 2015;131:e29–e322. - PubMed
-
- Halestrap AP. A pore way to die: the role of mitochondria in reperfusion injury and cardioprotection. Biochem Soc Trans. 2010;38:841–860. - PubMed
-
- Di Lisa F, Canton M, Carpi A, Kaludercic N, Menabò R, Menazza S, Semenzato M. Mitochondrial injury and protection in ischemic pre- and postconditioning. Antioxid Redox Signal. 2011;14:881–891. - PubMed
-
- Ong S-B, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

