Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population
- PMID: 29362361
- PMCID: PMC5780481
- DOI: 10.1038/s41467-017-01972-9
Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population
Abstract
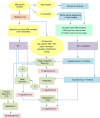
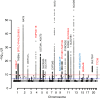
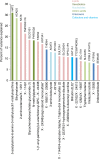
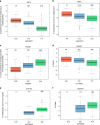
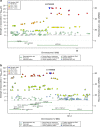
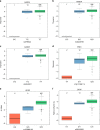
Metabolomics-genome-wide association studies (mGWAS) have uncovered many metabolic quantitative trait loci (mQTLs) influencing human metabolic individuality, though predominantly in European cohorts. By combining whole-exome sequencing with a high-resolution metabolomics profiling for a highly consanguineous Middle Eastern population, we discover 21 common variant and 12 functional rare variant mQTLs, of which 45% are novel altogether. We fine-map 10 common variant mQTLs to new metabolite ratio associations, and 11 common variant mQTLs to putative protein-altering variants. This is the first work to report common and rare variant mQTLs linked to diseases and/or pharmacological targets in a consanguineous Arab cohort, with wide implications for precision medicine in the Middle East.
Conflict of interest statement
R.P.M. is an employee of Metabolon Inc. The remaining authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

