Differential signaling pathways are initiated in macrophages during infection depending on the intracellular fate of Chlamydia spp
- PMID: 29363185
- PMCID: PMC6715151
- DOI: 10.1111/imcb.1033
Differential signaling pathways are initiated in macrophages during infection depending on the intracellular fate of Chlamydia spp
Abstract
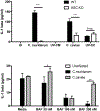
Chlamydia muridarum and Chlamydia caviae have equivalent growth rates in mouse epithelial cells but only C. muridarum replicates inside mouse macrophages, while C. caviae does not. Macrophages infected with C. muridarum or C. caviae were used to address the hypothesis that the early signaling pathways initiated during infection depend on the fate of chlamydiae in the host cell. Transmission electron microscopy of C. muridarum-infected macrophages showed intact chlamydial elementary bodies and reticulate bodies 2 h postinfection in compact vacuoles. Conversely, in macrophages infected with C. caviae, chlamydiae were observed in large phagocytic vacuoles. Furthermore, C. caviae infections failed to develop into inclusions or produce viable bacteria. Expression of proinflammatory cytokines TNFα, IL-1β and MMP13 was similar in C. caviae- or C. muridarum-infected macrophages at 3 h postinfection, indicating that chlamydial survival is not required for initiation of these responses. IL-1β secretion, dependent on inflammasome activation, occurred in C. caviae-infected macrophages despite no chlamydial growth. Conversely, IFNβ mRNA was observed only in C. muridarum- but not in C. caviae-infected macrophages. These data demonstrate that differential signaling events are initiated during a productive versus nonproductive chlamydial infection in a macrophage.
Keywords: Chlamydia; IFNβ; IL-1β; macrophages.
© 2017 Australasian Society for Immunology Inc.
Conflict of interest statement
CONFLICT OF INTEREST
All authors have no conflict of interests
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

