InfAcrOnt: calculating cross-ontology term similarities using information flow by a random walk
- PMID: 29363423
- PMCID: PMC5780854
- DOI: 10.1186/s12864-017-4338-6
InfAcrOnt: calculating cross-ontology term similarities using information flow by a random walk
Abstract
Background: Since the establishment of the first biomedical ontology Gene Ontology (GO), the number of biomedical ontology has increased dramatically. Nowadays over 300 ontologies have been built including extensively used Disease Ontology (DO) and Human Phenotype Ontology (HPO). Because of the advantage of identifying novel relationships between terms, calculating similarity between ontology terms is one of the major tasks in this research area. Though similarities between terms within each ontology have been studied with in silico methods, term similarities across different ontologies were not investigated as deeply. The latest method took advantage of gene functional interaction network (GFIN) to explore such inter-ontology similarities of terms. However, it only used gene interactions and failed to make full use of the connectivity among gene nodes of the network. In addition, all existent methods are particularly designed for GO and their performances on the extended ontology community remain unknown.
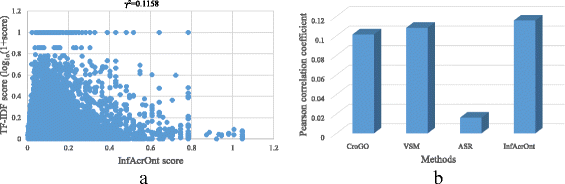
Results: We proposed a method InfAcrOnt to infer similarities between terms across ontologies utilizing the entire GFIN. InfAcrOnt builds a term-gene-gene network which comprised ontology annotations and GFIN, and acquires similarities between terms across ontologies through modeling the information flow within the network by random walk. In our benchmark experiments on sub-ontologies of GO, InfAcrOnt achieves a high average area under the receiver operating characteristic curve (AUC) (0.9322 and 0.9309) and low standard deviations (1.8746e-6 and 3.0977e-6) in both human and yeast benchmark datasets exhibiting superior performance. Meanwhile, comparisons of InfAcrOnt results and prior knowledge on pair-wise DO-HPO terms and pair-wise DO-GO terms show high correlations.
Conclusions: The experiment results show that InfAcrOnt significantly improves the performance of inferring similarities between terms across ontologies in benchmark set.
Keywords: Biomedical ontology; Information flow; Random walk; Term similarities.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Identifying term relations cross different gene ontology categories.BMC Bioinformatics. 2017 Dec 28;18(Suppl 16):573. doi: 10.1186/s12859-017-1959-3. BMC Bioinformatics. 2017. PMID: 29297309 Free PMC article.
-
Inferring gene ontologies from pairwise similarity data.Bioinformatics. 2014 Jun 15;30(12):i34-42. doi: 10.1093/bioinformatics/btu282. Bioinformatics. 2014. PMID: 24932003 Free PMC article.
-
Improving the measurement of semantic similarity by combining gene ontology and co-functional network: a random walk based approach.BMC Syst Biol. 2018 Mar 19;12(Suppl 2):18. doi: 10.1186/s12918-018-0539-0. BMC Syst Biol. 2018. PMID: 29560823 Free PMC article.
-
Ontology annotation: mapping genomic regions to biological function.Curr Opin Chem Biol. 2007 Feb;11(1):4-11. doi: 10.1016/j.cbpa.2006.11.039. Epub 2007 Jan 5. Curr Opin Chem Biol. 2007. PMID: 17208035 Review.
-
Ontology-driven approaches to analyzing data in functional genomics.Methods Mol Biol. 2006;316:67-86. doi: 10.1385/1-59259-964-8:67. Methods Mol Biol. 2006. PMID: 16671401 Review.
Cited by
-
Ultrasound Image Classification of Thyroid Nodules Based on Deep Learning.Front Oncol. 2022 Jul 15;12:905955. doi: 10.3389/fonc.2022.905955. eCollection 2022. Front Oncol. 2022. PMID: 35912199 Free PMC article.
-
Identification of Alzheimer's Disease-Related Genes Based on Data Integration Method.Front Genet. 2019 Jan 25;9:703. doi: 10.3389/fgene.2018.00703. eCollection 2018. Front Genet. 2019. PMID: 30740125 Free PMC article.
-
RF-PseU: A Random Forest Predictor for RNA Pseudouridine Sites.Front Bioeng Biotechnol. 2020 Feb 26;8:134. doi: 10.3389/fbioe.2020.00134. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32175316 Free PMC article.
-
Identification and Classification of Enhancers Using Dimension Reduction Technique and Recurrent Neural Network.Comput Math Methods Med. 2020 Oct 18;2020:8852258. doi: 10.1155/2020/8852258. eCollection 2020. Comput Math Methods Med. 2020. PMID: 33133227 Free PMC article.
-
eQTLMAPT: Fast and Accurate eQTL Mediation Analysis With Efficient Permutation Testing Approaches.Front Genet. 2020 Jan 9;10:1309. doi: 10.3389/fgene.2019.01309. eCollection 2019. Front Genet. 2020. PMID: 31998368 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

